File size: 7,362 Bytes
3fcb9c5 3fc9281 3fcb9c5 f67a52a 3fcb9c5 fb7aff2 3fcb9c5 0781fda f09ec43 0781fda c522049 f67a52a 9127336 f67a52a 44a02ef 6957904 0781fda 3fcb9c5 d9f7080 7f224e1 d9f7080 955192f 7f224e1 0781fda 7f224e1 2f5fb94 955192f 7f224e1 955192f 7f224e1 955192f d46f929 7f224e1 955192f 7f224e1 6dfc194 7f224e1 6dfc194 7f224e1 6dfc194 7f224e1 6dfc194 7f224e1 d46f929 7f224e1 a7890f1 7f224e1 a7890f1 7f224e1 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 |
---
# base_model: owkin/phikon
tags:
- feature-extraction
- image-classification
- timm
- biology
- cancer
- owkin
- histology
library_name: timm
model-index:
- name: owkin_pancancer
results:
- task:
type: image-classification
name: Image Classification
dataset:
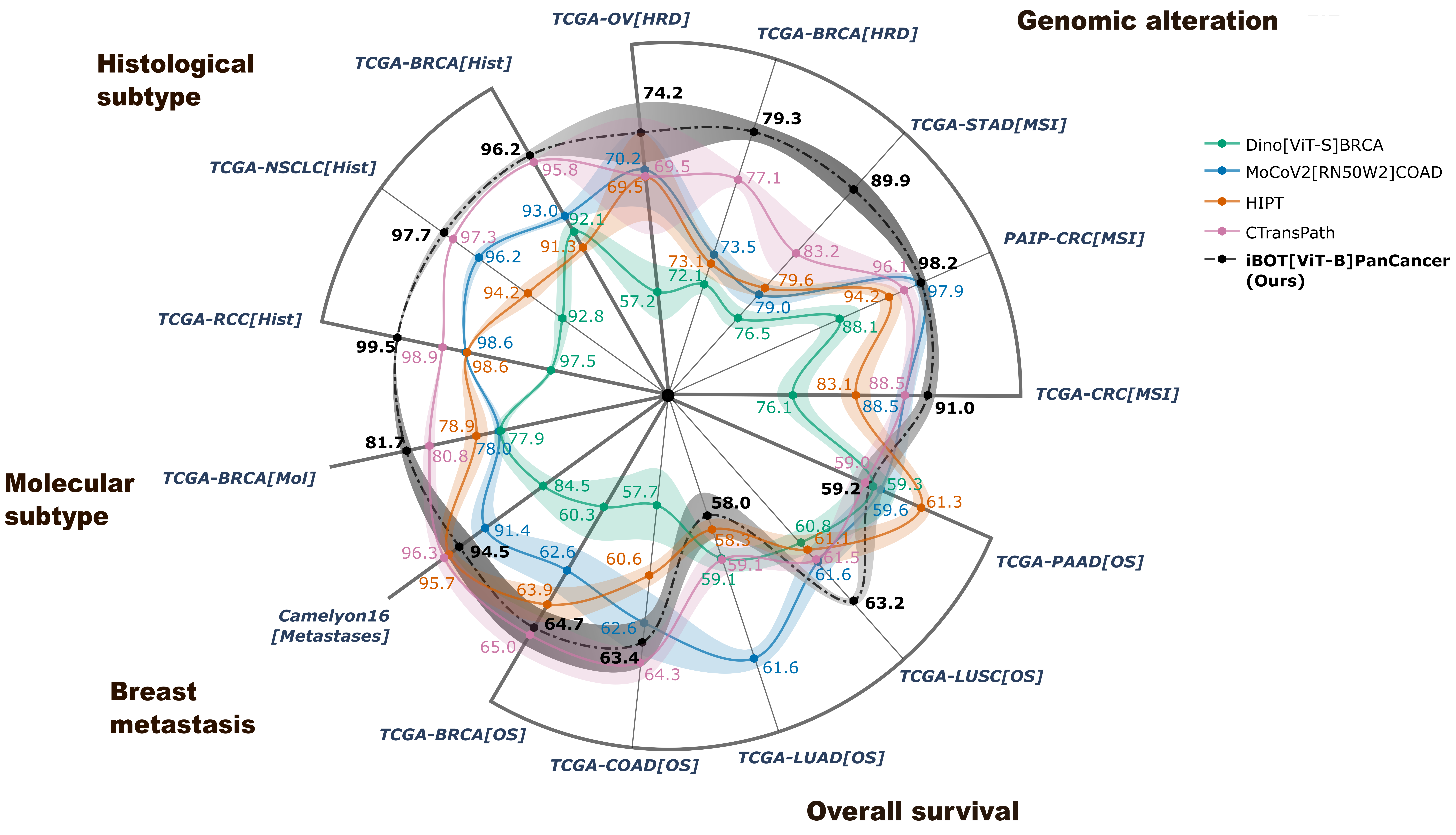
name: Camelyon16[Meta]
type: image-classification
metrics:
- type: accuracy
value: 94.5 ± 4.4
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[Hist]
type: image-classification
metrics:
- type: accuracy
value: 96.2 ± 3.3
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[HRD]
type: image-classification
metrics:
- type: accuracy
value: 79.3 ± 2.4
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[Mol]
type: image-classification
metrics:
- type: accuracy
value: 81.7 ± 1.6
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[OS]
type: image-classification
metrics:
- type: accuracy
value: 64.7 ± 5.7
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-CRC[MSI]
type: image-classification
metrics:
- type: accuracy
value: 91.0 ± 2.2
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-COAD[OS]
type: image-classification
metrics:
- type: accuracy
value: 63.4 ± 7.4
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-NSCLC[CType]
type: image-classification
metrics:
- type: accuracy
value: 97.7 ± 1.3
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-LUAD[OS]
type: image-classification
metrics:
- type: accuracy
value: 53.8 ± 4.5
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-LUSC[OS]
type: image-classification
metrics:
- type: accuracy
value: 62.2 ± 2.9
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-OV[HRD]
type: image-classification
metrics:
- type: accuracy
value: 74.2 ± 8.6
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-RCC[CType]
type: image-classification
metrics:
- type: accuracy
value: 99.5 ± 0.2
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-STAD[MSI]
type: image-classification
metrics:
- type: accuracy
value: 89.9 ± 3.9
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-PAAD[OS]
type: image-classification
metrics:
- type: accuracy
value: 59.2 ± 4.1
name: ROC AUC
verified: false
widget:
- src: https://github.com/owkin/HistoSSLscaling/raw/main/assets/example.tif
example_title: pancancer tile
co2_eq_emissions:
emissions: 14590
source: https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v2
training_type: pre-training
geographical_location: Jean Zay cluster, France (~40 gCO₂eq/kWh)
hardware_used: 32 V100 32Gb GPUs, 1216 GPU hours
license: other
license_name: owkin-non-commercial
license_link: https://github.com/owkin/HistoSSLscaling/blob/main/LICENSE.txt
pipeline_tag: feature-extraction
inference: false
datasets:
- owkin/camelyon16-features
- owkin/nct-crc-he
metrics:
- roc_auc
---
# Model card for vit_base_patch16_224.owkin_pancancer
A Vision Transformer (ViT) image classification model. \
Trained by Owkin on 40 million pan-cancer histology tiles from TCGA-COAD.
A version using the transformers library is also available here: https://huggingface.co/owkin/phikon
## Model Details
- **Model Type:** Feature backbone
- **Developed by**: Owkin
- **Funded by**: Owkin and IDRIS
- **Model Stats:**
- Params: 85.8M (base)
- Image size: 224 x 224 x 3
- Patch size: 16 x 16 x 3
- **Pre-training:**
- Dataset: Pancancer40M, created from [TCGA-COAD](https://portal.gdc.cancer.gov/repository?facetTab=cases&filters=%7B%22content%22%3A%5B%7B%22content%22%3A%7B%22field%22%3A%22cases.project.project_id%22%2C%22value%22%3A%5B%22TCGA-COAD%22%5D%7D%2C%22op%22%3A%22in%22%7D%2C%7B%22content%22%3A%7B%22field%22%3A%22files.experimental_strategy%22%2C%22value%22%3A%5B%22Diagnostic%20Slide%22%5D%7D%2C%22op%22%3A%22in%22%7D%5D%2C%22op%22%3A%22and%22%7D&searchTableTab=cases)
- Framework: [iBOT](https://github.com/bytedance/ibot), self-supervised, masked image modeling, self-distillation
- **Papers:**
- [Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling](https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v2)
- **Original:** https://github.com/owkin/HistoSSLscaling
- **License:** [Owkin non-commercial license](https://github.com/owkin/HistoSSLscaling/blob/main/LICENSE.txt)
## Model Usage
### Image Embeddings
```python
from urllib.request import urlopen
from PIL import Image
import timm
# get example histology image
img = Image.open(
urlopen(
"https://github.com/owkin/HistoSSLscaling/raw/main/assets/example.tif"
)
)
# load model from the hub
model = timm.create_model(
model_name="hf-hub:1aurent/vit_base_patch16_224.owkin_pancancer",
pretrained=True,
).eval()
# get model specific transforms (normalization, resize)
data_config = timm.data.resolve_model_data_config(model)
transforms = timm.data.create_transform(**data_config, is_training=False)
data = transforms(img).unsqueeze(0) # input is a (batch_size, num_channels, img_size, img_size) shaped tensor
output = model(data) # output is a (batch_size, num_features) shaped tensor
```
## Citation
```bibtex
@article{Filiot2023.07.21.23292757,
author = {Alexandre Filiot and Ridouane Ghermi and Antoine Olivier and Paul Jacob and Lucas Fidon and Alice Mac Kain and Charlie Saillard and Jean-Baptiste Schiratti},
title = {Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling},
elocation-id = {2023.07.21.23292757},
year = {2023},
doi = {10.1101/2023.07.21.23292757},
publisher = {Cold Spring Harbor Laboratory Press},
url = {https://www.medrxiv.org/content/early/2023/09/14/2023.07.21.23292757},
eprint = {https://www.medrxiv.org/content/early/2023/09/14/2023.07.21.23292757.full.pdf},
journal = {medRxiv}
}
``` |