|
--- |
|
language: rna |
|
tags: |
|
- Biology |
|
- RNA |
|
license: |
|
- agpl-3.0 |
|
size_categories: |
|
- 10M<n<100M |
|
source_datasets: |
|
- multimolecule/5srrnadb |
|
- multimolecule/crw |
|
- multimolecule/dictybase |
|
- multimolecule/ena |
|
- multimolecule/ensembl |
|
- multimolecule/ensembl_fungi |
|
- multimolecule/ensembl_gencode |
|
- multimolecule/ensembl_metazoa |
|
- multimolecule/ensembl_plants |
|
- multimolecule/ensembl_protists |
|
- multimolecule/evlncrnas |
|
- multimolecule/expression_atlas |
|
- multimolecule/flybase |
|
- multimolecule/genecards |
|
- multimolecule/greengenes |
|
- multimolecule/gtrnadb |
|
- multimolecule/hgnc |
|
- multimolecule/intact |
|
- multimolecule/lncbase |
|
- multimolecule/lncbook |
|
- multimolecule/lncipedia |
|
- multimolecule/lncrnadb |
|
- multimolecule/malacards |
|
- multimolecule/mgi |
|
- multimolecule/mgnify |
|
- multimolecule/mirbase |
|
- multimolecule/mirgenedb |
|
- multimolecule/modomics |
|
- multimolecule/noncode |
|
- multimolecule/pdbe |
|
- multimolecule/pirbase |
|
- multimolecule/plncdb |
|
- multimolecule/pombase |
|
- multimolecule/psicquic |
|
- multimolecule/rdp |
|
- multimolecule/refseq |
|
- multimolecule/rfam |
|
- multimolecule/rgd |
|
- multimolecule/ribocentre |
|
- multimolecule/ribovision |
|
- multimolecule/sgd |
|
- multimolecule/silva |
|
- multimolecule/snodb |
|
- multimolecule/snopy |
|
- multimolecule/snorna_database |
|
- multimolecule/srpdb |
|
- multimolecule/tair |
|
- multimolecule/tarbase |
|
- multimolecule/tmrna_website |
|
- multimolecule/wormbase |
|
- multimolecule/zfin |
|
- multimolecule/zwd |
|
task_categories: |
|
- text-generation |
|
- fill-mask |
|
task_ids: |
|
- language-modeling |
|
- masked-language-modeling |
|
pretty_name: RNAcentral |
|
library_name: multimolecule |
|
--- |
|
|
|
# RNAcentral |
|
|
|
 |
|
|
|
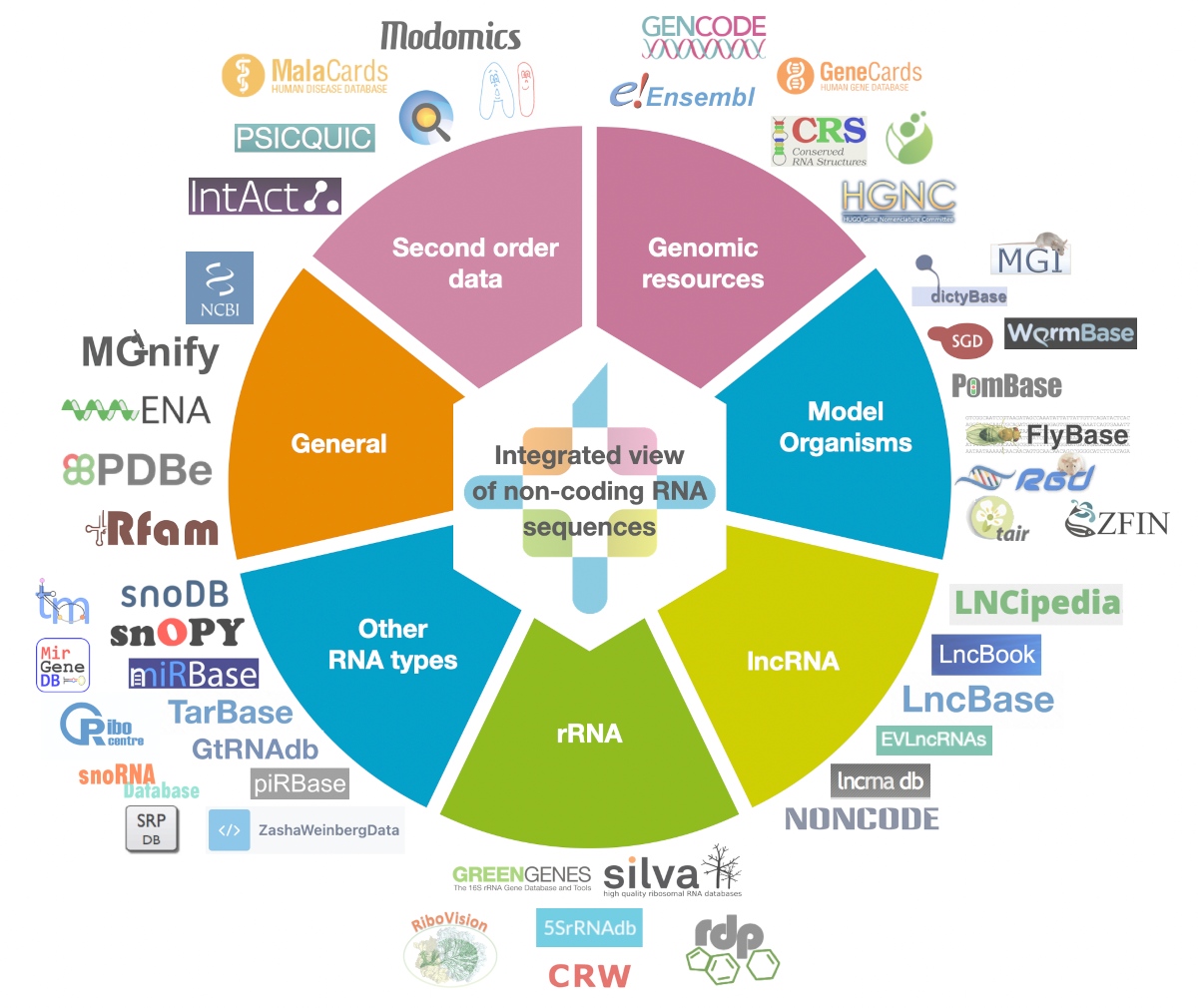
RNAcentral is a free, public resource that offers integrated access to a comprehensive and up-to-date set of non-coding RNA sequences provided by a collaborating group of [Expert Databases](https://rnacentral.org/expert-databases) representing a broad range of organisms and RNA types. |
|
|
|
The development of RNAcentral is coordinated by [European Bioinformatics Institute](http://www.ebi.ac.uk/) and is supported by [Wellcome](https://wellcome.ac.uk/). Initial funding was provided by [BBSRC](https://bbsrc.ukri.org/). |
|
|
|
## Disclaimer |
|
|
|
This is an UNOFFICIAL release of the [RNAcentral](https://rnacentral.org) by The RNAcentral Consortium. |
|
|
|
**The team releasing RNAcentral did not write this dataset card for this dataset so this dataset card has been written by the MultiMolecule team.** |
|
|
|
## Dataset Description |
|
|
|
- **Homepage**: https://rnacentral.org |
|
- **Documentation**: https://multimolecule.danling.org/datasets/rnacentral |
|
- **datasets**: https://huggingface.co/datasets/multimolecule/rnacentral |
|
- **Point of Contact**: [Blake Sweeney](https://www.ebi.ac.uk/people/person/blake-sweeney/) |
|
|
|
## Derived Datasets |
|
|
|
- [rnacentral](https://huggingface.co/datasets/multimolecule/rnacentral): RNAcentral dataset. |
|
- [rnacentral-secondary_structure](https://huggingface.co/datasets/multimolecule/rnacentral-secondary_structure): RNAcentral subset with secondary structure annotations. |
|
- [rnacentral-modifications](https://huggingface.co/datasets/multimolecule/rnacentral-modifications): RNAcentral subset with modifications annotations. |
|
|
|
## License |
|
|
|
This dataset is licensed under the [AGPL-3.0 License](https://www.gnu.org/licenses/agpl-3.0.html). |
|
|
|
```spdx |
|
SPDX-License-Identifier: AGPL-3.0-or-later |
|
``` |
|
|
|
> [!TIP] |
|
> The original RNAcentral dataset is licensed under the CC0 1.0 Universal (CC0 1.0) Public Domain Dedication. |
|
> |
|
> This dataset is a derivative of the original RNAcentral dataset and uses a different license. |
|
|
|
## Citation |
|
|
|
```bibtex |
|
@article{rnacentral2021, |
|
author = {{RNAcentral Consortium}}, |
|
doi = {https://doi.org/10.1093/nar/gkaa921}, |
|
journal = {Nucleic Acids Research}, |
|
month = jan, |
|
number = {D1}, |
|
pages = {D212--D220}, |
|
publisher = {Oxford University Press (OUP)}, |
|
title = {{RNAcentral} 2021: secondary structure integration, improved sequence search and new member databases}, |
|
url = {https://academic.oup.com/nar/article/49/D1/D212/5940500}, |
|
volume = 49, |
|
year = 2021 |
|
} |
|
|
|
@article{sweeney2020exploring, |
|
author = {Sweeney, Blake A. and Tagmazian, Arina A. and Ribas, Carlos E. and Finn, Robert D. and Bateman, Alex and Petrov, Anton I.}, |
|
doi = {https://doi.org/10.1002/cpbi.104}, |
|
eprint = {https://currentprotocols.onlinelibrary.wiley.com/doi/pdf/10.1002/cpbi.104}, |
|
journal = {Current Protocols in Bioinformatics}, |
|
keywords = {Galaxy, ncRNA, non-coding RNA, RNAcentral, RNA-seq}, |
|
number = {1}, |
|
pages = {e104}, |
|
title = {Exploring Non-Coding RNAs in RNAcentral}, |
|
url = {https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/cpbi.104}, |
|
volume = 71, |
|
year = 2020 |
|
} |
|
|
|
@article{rnacentral2019, |
|
author = {{The RNAcentral Consortium}}, |
|
doi = {https://doi.org/10.1093/nar/gky1034}, |
|
journal = {Nucleic Acids Research}, |
|
month = jan, |
|
number = {D1}, |
|
pages = {D221--D229}, |
|
publisher = {Oxford University Press (OUP)}, |
|
title = {{RNAcentral}: a hub of information for non-coding {RNA} sequences}, |
|
url = {https://academic.oup.com/nar/article/47/D1/D221/5160993}, |
|
volume = 47, |
|
year = 2019 |
|
} |
|
|
|
@article{rnacentral2017, |
|
author = {{The RNAcentral Consortium} and Petrov, Anton I and Kay, Simon J E and Kalvari, Ioanna and Howe, Kevin L and Gray, Kristian A and Bruford, Elspeth A and Kersey, Paul J and Cochrane, Guy and Finn, Robert D and Bateman, Alex and Kozomara, Ana and Griffiths-Jones, Sam and Frankish, Adam and Zwieb, Christian W and Lau, Britney Y and Williams, Kelly P and Chan, Patricia Pand Lowe, Todd M and Cannone, Jamie J and Gutell, Robin and Machnicka, Magdalena A and Bujnicki, Janusz M and Yoshihama, Maki and Kenmochi, Naoya and Chai, Benli and Cole, James R and Szymanski, Maciej and Karlowski, Wojciech M and Wood, Valerie and Huala, Eva and Berardini, Tanya Z and Zhao, Yi and Chen, Runsheng and Zhu, Weimin and Paraskevopoulou, Maria D and Vlachos, Ioannis S and Hatzigeorgiou, Artemis G and Ma, Lina and Zhang, Zhang and Puetz, Joern and Stadler, Peter F and McDonald, Daniel and Basu, Siddhartha and Fey, Petra and Engel, Stacia R and Cherry, J Michael and Volders, Pieter-Jan and Mestdagh, Pieter and Wower, Jacek and Clark, Michael B and Quek, Xiu Cheng and Dinger, Marcel E}, |
|
doi = {https://doi.org/10.1093/nar/gkw1008}, |
|
journal = {Nucleic Acids Research}, |
|
month = jan, |
|
number = {D1}, |
|
pages = {D128--D134}, |
|
publisher = {Oxford University Press (OUP)}, |
|
title = {{RNAcentral}: a comprehensive database of non-coding {RNA} sequences}, |
|
url = {https://academic.oup.com/nar/article/45/D1/D128/2333921}, |
|
volume = 45, |
|
year = 2017 |
|
} |
|
|
|
@article{rnacentral2015, |
|
author = {{RNAcentral Consortium} and Petrov, Anton I and Kay, Simon J E and Gibson, Richard and Kulesha, Eugene and Staines, Dan and Bruford, Elspeth A and Wright, Mathew W and Burge, Sarah and Finn, Robert D and Kersey, Paul J and Cochrane, Guy and Bateman, Alex and Griffiths-Jones, Sam and Harrow, Jennifer and Chan, Patricia P and Lowe, Todd M and Zwieb, Christian W and Wower, Jacek and Williams, Kelly P and Hudson, Corey M and Gutell, Robin and Clark, Michael B and Dinger, Marcel and Quek, Xiu Cheng and Bujnicki, Janusz M and Chua, Nam-Hai and Liu, Jun and Wang, Huan and Skogerb{\o}, Geir and Zhao, Yi and Chen, Runsheng and Zhu, Weimin and Cole, James R and Chai, Benli and Huang, Hsien-Da and Huang, His-Yuan and Cherry, J Michael and Hatzigeorgiou, Artemis and Pruitt, Kim D}, |
|
doi = {https://doi.org/10.1093/nar/gku991}, |
|
journal = {Nucleic Acids Research}, |
|
month = jan, |
|
number = {Database issue}, |
|
pages = {D123--D129}, |
|
title = {{RNAcentral}: an international database of {ncRNA} sequences}, |
|
url = {https://academic.oup.com/nar/article/43/D1/D123/2439941}, |
|
volume = 43, |
|
year = 2015 |
|
} |
|
|
|
@article{bateman2011rnacentral, |
|
author = {Bateman, Alex and Agrawal, Shipra and Birney, Ewan and Bruford, Elspeth A and Bujnicki, Janusz M and Cochrane, Guy and Cole, James R and Dinger, Marcel E and Enright, Anton J and Gardner, Paul P and Gautheret, Daniel and Griffiths-Jones, Sam and Harrow, Jen and Herrero, Javier and Holmes, Ian H and Huang, Hsien-Da and Kelly, Krystyna A and Kersey, Paul and Kozomara, Ana and Lowe, Todd M and Marz, Manja and Moxon, Simon andPruitt, Kim D and Samuelsson, Tore and Stadler, Peter F and Vilella, Albert J and Vogel, Jan-Hinnerk and Williams, Kelly P and Wright, Mathew W and Zwieb, Christian}, |
|
doi = {https://doi.org/10.1261/rna.2750811}, |
|
journal = {RNA}, |
|
month = nov, |
|
number = 11, |
|
pages = {1941--1946}, |
|
publisher = {Cold Spring Harbor Laboratory}, |
|
title = {{RNAcentral}: A vision for an international database of {RNA} sequences}, |
|
url = {https://rnajournal.cshlp.org/content/17/11/1941.long}, |
|
volume = 17, |
|
year = 2011 |
|
} |
|
``` |
|
|