prompt
stringlengths 19
1.03M
| completion
stringlengths 4
2.12k
| api
stringlengths 8
90
|
|---|---|---|
# _____ _ _ _ ____ _ _ _
# |_ _| |__ (_)___ (_)___ | _ \(_)_ __ _ _| | __ _| |_ ___
# | | | '_ \| / __| | / __| | |_) | | '_ \| | | | |/ _` | __/ _ \
# | | | | | | \__ \ | \__ \ | __/| | |_) | |_| | | (_| | || __/
# |_| |_| |_|_|___/ |_|___/ |_| |_| .__/ \__,_|_|\__,_|\__\___|
# There are many frameworks like it, |_| but this one is mine. --MikeL
# IMPORTANT THINGS TO REMEMBER:
# 1. Make life easy by filling your ~/.bash_profile with aliases.
# 2. Make life easy by filling /usr/local/sbin with helper scripts.
# 3. pip puts things ~here: /usr/local/lib/python3.5/dist-packages/
# 4. oauth2client is deprecated. Replace it soon.
import sys
import os
import gspread
import httplib2
from datetime import date, datetime, timedelta
from inspect import getfile, currentframe, getouterframes, stack
from collections import defaultdict
import pytz
from oauth2client.client import OAuth2WebServerFlow
from oauth2client import file, tools
import pandas as pd
from time import sleep, gmtime, strftime
from pyfiglet import figlet_format
from colorama import Fore
from logzero import logger, setup_logger
filename = "oauth.dat"
client_id = "769904540573-knscs3mhvd56odnf7i8h3al13kiqulft.apps.googleusercontent.com"
client_secret = "<KEY>"
# To count how frequently functions have been called.
counters = defaultdict(int)
def return_error():
"""Return error tuple and stop execution. Useful on naked exceptions."""
e = sys.exc_info()
print(e)
raise SystemExit()
def pipulate(tab, rows, cols, columns=None):
"""All that pipulate really is"""
row1, row2 = rows
col1, col2 = cols
col1, col2 = aa(col1), aa(col2)
cl = tab.range(row1, col1, row2, col2)
list_of_tuples = cl_to_tuples(cl)
if not columns:
columns = tab.range(row1, col1, row1, col2)
columns = [cc(x.col) for x in columns]
df = | pd.DataFrame(list_of_tuples, columns=columns) | pandas.DataFrame |
# -*- coding:utf-8 -*-
# Author: <NAME>
# Data: 2/20/2018
# Describe: Build a dictionary based on my own needs.
import os.path
import re
import pandas as pd
import translate as tl
class Dictionary(object):
def __init__(self, dic_name):
if not os.path.isfile(dic_name):
open(dic_name, 'a').close()
self.total_words = 0
self.dic_name = dic_name
self.columns = ['英', '汉']
self.pronounce = True
self.nostorage = False
try:
self.dic = pd.read_csv(dic_name)
except pd.io.common.EmptyDataError:
self.dic = | pd.DataFrame(columns=self.columns, dtype=str) | pandas.DataFrame |
import numpy as np
import pandas as pd
import pickle
import time
import random
import os
from sklearn import linear_model, model_selection, ensemble
from sklearn.svm import SVC
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.base import clone
from sklearn import metrics
from sklearn.model_selection import cross_validate, train_test_split, StratifiedKFold
import sklearn.metrics as m
from joblib import Parallel, delayed
from sklearn.base import clone
from sklearn.utils import shuffle, resample
type_='marker'
basename = type_+'_features_expired_prediction_'
dir_ = '../../data/'
t0_all=time.time()
seed = 42
np.random.seed(seed)
max_depth = 1
C=1
tol=1e-3
min_samples_leaf=2
min_samples_split=2
n_estimators=100
models = {
"Logistic Regression" : linear_model.LogisticRegression(
C=C,
penalty='l1',
solver="liblinear",
tol=tol,
random_state=seed)
}
classification_metrics = ['roc_auc']
cv_split = 10
test_size = 0.15
n_jobs = 25
nboot=200
X_all_proteins = pd.read_csv(dir_+'integrated_X_raw_all_proteins.csv',index_col=0)
proteins_no_immunoglobulins = pickle.load(open(dir_+'proteins_no_immunoglobulins.pkl','rb'))
X_all_proteins = X_all_proteins.loc[:,proteins_no_immunoglobulins]
joined = pd.read_csv(dir_+'mortality_X_y.csv',index_col=0)
X_all_clinical = pd.read_csv(dir_+'integrated_X_clinical_and_cohort_covariates.csv',index_col=0)
Y_pgd = pd.read_csv(dir_+'integrated_pgd_y.csv',index_col=0,header=None)
Y_pgd.columns = ['PGD']
X_all_clinical = X_all_clinical.join(Y_pgd)
Y_mortality = joined[['expired']]
Y_mortality.index.name=''
X_all_clinical = X_all_clinical.join(Y_mortality)
Y_lvad = joined[['Mechanical_Support_Y']]
Y_lvad.index.name=''
Y_survival = (joined[['expired']]==0).astype(int)
Y_survival.columns = ['Survival']
Y_survival.index.name=''
X_all_clinical = X_all_clinical.join(Y_survival)
idmap_sub = pd.read_csv(dir_+'protein_gene_map_full.csv')[['Protein','Gene_name']].dropna()
cov_df = X_all_clinical.loc[:,['Cohort_Columbia','Cohort_Cedar']].copy().astype(int)
all_cov_df = cov_df.copy()
all_cov_df.loc[:,'Cohort_Paris'] = (
(all_cov_df['Cohort_Columbia'] +
all_cov_df['Cohort_Cedar'])==0).astype(int)
params = {'Y' : Y_survival, 'cv_split' : cv_split,
'metrics' : classification_metrics, 'n_jobs' : 1,
'test_size' : test_size,
'retrained_models' : True, 'patient_level_predictions' : True}
def permute(Y,seed=42):
"""
shuffle sample values
Parameters:
----------
Y : pandas series
Index of samples and values are their class labels
seed : int
Random seed for shuffling
Returns:
------
arr_shuffle: pandas series
A shuffled Y
"""
arr = shuffle(Y.values,random_state=seed)
arr_shuffle = (pd.Series(arr.reshape(1,-1)[0],index=Y.index))
return arr_shuffle
def observed_val(X,Y,models,metrics=['roc_auc'],cv_split=10,seed=42,test_size=0.15,return_train_score=False,n_jobs=1,retrained_models=False,patient_level_predictions=False,return_estimator=True):
# make sure given metrics are in list and not one metric is given as a string
if type(metrics)!=list:
metrics = [metrics]
# 1/ reindex
X = X.loc[Y.index]
dfs = []
model_retrained_fits = {}
model_confs = []
#iterate through model dictionary
for name,mod in models.items():
# /2 generate model parameters and fold scores with cv splitter
fit = clone(mod).fit(X,Y.values.reshape(1,-1)[0])
conf = pd.DataFrame({'y_true' : Y.values.reshape(1,-1)[0],'y_pred' : fit.predict(X),'bootstrap' : 'observed','model' : np.repeat(name,len(Y.index))},index=Y.index)
model_confs.append(conf)
#do prediction for each metric
tmp = pd.DataFrame({'model' : name,'bootstrap' : 'observed'},index=[0])
for metric in metrics:
tmp[metric] = m.SCORERS[metric](fit,X,Y)
model_retrained_fits[name] = fit
dfs.append(tmp)
return pd.concat(dfs).reset_index(drop=True), model_retrained_fits, pd.concat(model_confs)
def resample_observed_val(X,Y,models,metrics=['roc_auc'],cv_split=10,seed=42,test_size=0.15,return_train_score=False,n_jobs=1,retrained_models=False,patient_level_predictions=False,return_estimator=True):
# make sure given metrics are in list and not one metric is given as a string
if type(metrics)!=list:
metrics = [metrics]
# 1/ reindex
X = X.loc[Y.index]
Y_resample = resample(Y,random_state=seed)
X = X.loc[Y_resample.index]
Y = Y_resample.copy()
dfs = []
model_retrained_fits = {}
model_confs = []
#iterate through model dictionary
for name,mod in models.items():
# /2 generate model parameters and fold scores with cv splitter
fit = clone(mod).fit(X,Y.values.reshape(1,-1)[0])
conf = pd.DataFrame({'y_true' : Y.values.reshape(1,-1)[0],'y_pred' : fit.predict(X),'bootstrap' : 'observed','model' : np.repeat(name,len(Y.index))},index=Y.index)
model_confs.append(conf)
#do prediction for each metric
tmp = pd.DataFrame({'model' : name,'bootstrap' : 'observed'},index=[0])
for metric in metrics:
tmp[metric] = m.SCORERS[metric](fit,X,Y)
model_retrained_fits[name] = fit
dfs.append(tmp)
return pd.concat(dfs).reset_index(drop=True), model_retrained_fits, pd.concat(model_confs)
def permuted_observed_val(X,Y,models,metrics=['roc_auc'],cv_split=10,seed=42,test_size=0.15,return_train_score=False,n_jobs=1,retrained_models=False,patient_level_predictions=False,return_estimator=True):
# make sure given metrics are in list and not one metric is given as a string
if type(metrics)!=list:
metrics = [metrics]
# 1/ reindex
X = X.loc[Y.index]
Y_shuffle = permute(Y,seed=seed)
X = X.loc[Y_shuffle.index]
Y = Y_shuffle.copy()
dfs = []
model_retrained_fits = {}
model_confs = []
#iterate through model dictionary
for name,mod in models.items():
# /2 generate model parameters and fold scores with cv splitter
fit = clone(mod).fit(X,Y.values.reshape(1,-1)[0])
conf = pd.DataFrame({'y_true' : Y.values.reshape(1,-1)[0],'y_pred' : fit.predict(X),'bootstrap' : 'observed','model' : np.repeat(name,len(Y.index))},index=Y.index)
model_confs.append(conf)
#do prediction for each metric
tmp = pd.DataFrame({'model' : name,'bootstrap' : 'observed'},index=[0])
for metric in metrics:
tmp[metric] = m.SCORERS[metric](fit,X,Y)
model_retrained_fits[name] = fit
dfs.append(tmp)
return pd.concat(dfs).reset_index(drop=True), model_retrained_fits, pd.concat(model_confs)
def train_test_val_top_fold_01_within(X,Y,models,metrics=['roc_auc'],cv_split=10,seed=42,test_size=0.15,return_train_score=True,n_jobs=1,retrained_models=False,patient_level_predictions=False,return_estimator=True):
# make sure given metrics are in list and not one metric given as a string
if type(metrics)!=list:
metrics = [metrics]
# 1/ train and test split
X = X.loc[Y.index]
X_train, X_test, y_train, y_test = train_test_split(X,Y,
test_size=test_size,
random_state=seed,
stratify=Y,
shuffle=True)
X_train = X_train.apply(lambda x : (x - min(x))/(max(x) - min(x)),axis=0)
X_test = X_test.apply(lambda x : (x - min(x))/(max(x) - min(x)),axis=0)
X_train[X_train.isna()]=0
X_test[X_test.isna()]=0
#define K fold splitter
cv = StratifiedKFold(n_splits=cv_split,random_state=seed,shuffle=True)
#Instantiate lists to collect prediction and model results
dfs = []
model_retrained_fits = {}
model_confs = []
#iterate through model dictionary
for name,mod in models.items():
# /2 generate model parameters and fold scores with cv splitter
fit = cross_validate(clone(mod),X_train,y_train.values.reshape(1,-1)[0],cv=cv,scoring=metrics,
n_jobs=n_jobs,return_train_score=return_train_score,
return_estimator=return_estimator)
tmp = pd.DataFrame({'fold' : range(cv_split),
'model' : name},
index=range(cv_split))
#populate scores in dataframe
cols = [k for k in fit.keys() if (k.find('test')+k.find('train'))==-1]
for col in cols:
tmp[col] = fit[col]
# /3 Identify best performing model
top_fold = np.where(fit['test_roc_auc']==fit['test_roc_auc'].max())[0][0]
keys = [x for x in fit.keys()]
vals = [fit[x][top_fold] for x in keys]
top_model_key_vals = {}
for i in range(len(vals)):
top_model_key_vals[keys[i]] = vals[i]
#4/ train models on training set
# also get sample level predictions
f = top_model_key_vals['estimator']
fitted = clone(f).fit(X_train,y_train.values.reshape(1,-1)[0])
conf = pd.DataFrame({'y_true' : y_test.values.reshape(1,-1)[0],
'y_pred' : fitted.predict(X_test),
'y_proba' : fitted.predict_proba(X_test)[:,1],
'bootstrap' : np.repeat(seed,len(y_test.index)),
'model' : np.repeat(name,len(y_test.index))},
index=y_test.index)
model_confs.append(conf)
#do prediction for each metric
for metric in metrics:
tmp['validation_'+metric] = m.SCORERS[metric](fitted,X_test,y_test)
model_retrained_fits[name] = fitted
dfs.append(tmp.query('fold==@top_fold').drop('fold',1))
return pd.concat(dfs,sort=True).reset_index(drop=True), model_retrained_fits, pd.concat(model_confs,sort=True)
def permuted_train_test_val_top_fold_01_within(X,Y,models,metrics=['roc_auc'],cv_split=10,seed=42,test_size=0.15,return_train_score=True,n_jobs=1,retrained_models=False,patient_level_predictions=False,return_estimator=True):
X = X.loc[Y.index]
Y_shuffle = permute(Y,seed=seed)
X_shuffle = X.loc[Y_shuffle.index]
# make sure given metrics are in list and not one metric given as a string
if type(metrics)!=list:
metrics = [metrics]
# 1/ train and test split
X_train, X_test, y_train, y_test = train_test_split(X_shuffle,Y_shuffle,
test_size=test_size,
random_state=seed,
stratify=Y,
shuffle=True)
X_train = X_train.apply(lambda x : (x - min(x))/(max(x) - min(x)),axis=0)
X_test = X_test.apply(lambda x : (x - min(x))/(max(x) - min(x)),axis=0)
X_train[X_train.isna()]=0
X_test[X_test.isna()]=0
#define K fold splitter
cv = StratifiedKFold(n_splits=cv_split,random_state=seed,shuffle=True)
#Instantiate lists to collect prediction and model results
dfs = []
model_retrained_fits = {}
model_confs = []
#iterate through model dictionary
for name,mod in models.items():
# /2 generate model parameters and fold scores with cv splitter
fit = cross_validate(clone(mod),X_train,y_train.values.reshape(1,-1)[0],cv=cv,scoring=metrics,
n_jobs=n_jobs,return_train_score=return_train_score,
return_estimator=return_estimator)
tmp = pd.DataFrame({'fold' : range(cv_split),
'model' : name},
index=range(cv_split))
#populate scores in dataframe
cols = [k for k in fit.keys() if (k.find('test')+k.find('train'))==-1]
for col in cols:
tmp[col] = fit[col]
# /3 Identify best performing model
top_fold = np.where(fit['test_roc_auc']==fit['test_roc_auc'].max())[0][0]
keys = [x for x in fit.keys()]
vals = [fit[x][top_fold] for x in keys]
top_model_key_vals = {}
for i in range(len(vals)):
top_model_key_vals[keys[i]] = vals[i]
#4/ train models on training set
# also get sample level predictions
f = top_model_key_vals['estimator']
fitted = clone(f).fit(X_train,y_train.values.reshape(1,-1)[0])
conf = pd.DataFrame({'y_true' : y_test.values.reshape(1,-1)[0],
'y_pred' : fitted.predict(X_test),
'y_proba' : fitted.predict_proba(X_test)[:,1],
'bootstrap' : np.repeat(seed,len(y_test.index)),
'model' : np.repeat(name,len(y_test.index))},
index=y_test.index)
model_confs.append(conf)
#do prediction for each metric
for metric in metrics:
tmp['validation_'+metric] = m.SCORERS[metric](fitted,X_test,y_test)
model_retrained_fits[name] = fitted
dfs.append(tmp.query('fold==@top_fold').drop('fold',1))
return pd.concat(dfs,sort=True).reset_index(drop=True), model_retrained_fits, pd.concat(model_confs,sort=True)
def bootstrap_of_fcn(func=None,params={},n_jobs=4,nboot=2):
if func==None:
return "Need fcn to bootstrap"
parallel = Parallel(n_jobs=n_jobs)
return parallel(
delayed(func)(
seed=k,**params)
for k in range(nboot))
def get_performance(lst):
perf = (pd.
concat(lst,keys=range(len(lst))).
reset_index(level=1,drop=True).
rename_axis('bootstrap').
reset_index()
)
return perf
def model_feature_importances(boot_mods):
dfs = []
X = params['X'].copy()
X.loc[:,'Intercept'] = 0
for i in range(len(boot_mods)):
for j in boot_mods[i].keys():
mod = boot_mods[i][j]
coef = []
try:
coef.extend([i for i in mod.feature_importances_])
except:
coef.extend([i for i in mod.coef_[0]])
coef.extend(mod.intercept_)
fs = []
fs.extend(X.columns.values)
df = pd.DataFrame({
'Feature' : fs,
'Gene_name' : (X.T.
join(idmap_sub.
set_index('Protein'),how='left').
Gene_name.values),
'Importance' : coef,
'Model' : j,
'Bootstrap' : i
})
dfs.append(df)
return pd.concat(dfs,sort=True)
def patient_predictions(lst):
col = pd.concat(lst).index.name
dat = \
(pd.
concat(
lst
).
reset_index().
rename(columns={col : 'Sample'}).
set_index('Sample').
join(all_cov_df).
reset_index().
melt(id_vars=['Sample','bootstrap','model','y_true','y_pred','y_proba'],
var_name='cohort',value_name='mem')
)
dat.cohort = dat.cohort.str.split('_').apply(lambda x : x[1])
dat = dat[dat.mem==1].drop('mem',1).reset_index(drop=True)
return dat
def get_performance(lst):
perf = (pd.
concat(lst,keys=range(len(lst))).
reset_index(level=1,drop=True).
rename_axis('bootstrap').
reset_index()
)
return perf
def model_feature_importances(boot_mods):
dfs = []
X = params['X'].copy()
X.loc[:,'Intercept'] = 0
for i in range(len(boot_mods)):
for j in boot_mods[i].keys():
mod = boot_mods[i][j]
coef = []
try:
coef.extend([i for i in mod.feature_importances_])
except:
coef.extend([i for i in mod.coef_[0]])
coef.extend(mod.intercept_)
fs = []
fs.extend(X.columns.values)
df = pd.DataFrame({
'Feature' : fs,
'Gene_name' : (X.T.
join(idmap_sub.
set_index('Protein'),how='left').
Gene_name.values),
'Importance' : coef,
'Model' : j,
'Bootstrap' : i
})
dfs.append(df)
return pd.concat(dfs,sort=True)
def patient_predictions(lst):
col = pd.concat(lst).index.name
dat = \
(pd.
concat(
lst
).
reset_index().
rename(columns={col : 'Sample'}).
set_index('Sample').
join(all_cov_df).
reset_index().
melt(id_vars=['Sample','bootstrap','model','y_true','y_pred','y_proba'],
var_name='cohort',value_name='mem')
)
dat.cohort = dat.cohort.str.split('_').apply(lambda x : x[1])
dat = dat[dat.mem==1].drop('mem',1).reset_index(drop=True)
return dat
import itertools
clin_combos = [[list(i) for i in itertools.combinations(
np.intersect1d(
X_all_clinical.columns.values,
X_all_clinical.columns.values),r)
] for r in np.arange(1,2)]
prot_combos = [[list(i) for i in itertools.combinations(
np.intersect1d(
X_all_proteins.columns.values,
X_all_proteins.columns.values),r)
] for r in np.arange(1,2)]
all_clin_1 = list(np.concatenate(list(itertools.chain(*clin_combos))))
print(len(all_clin_1))
all_prot_1 = list(np.concatenate(list(itertools.chain(*prot_combos))))
print(len(all_prot_1))
all_clin_1_and_prot_1 = list(
itertools.chain(*[all_clin_1,all_prot_1])
)
print(len(all_clin_1_and_prot_1))
all_clin_1_prot_1 = list(
itertools.chain(*
[[list(itertools.chain(*[[x],[y]])) for x in all_prot_1] for y in all_clin_1]
)
)
print(len(all_clin_1_prot_1))
all_clin_1_prot_1_and_clin_1_and_prot_1 = list(
itertools.chain(*[all_clin_1,all_prot_1,all_clin_1_prot_1])
)
print(len(all_clin_1_prot_1_and_clin_1_and_prot_1))
all_clin_2 = [list(i) for i in itertools.combinations(all_clin_1,2)]
print(len(all_clin_2))
all_prot_2 = [list(i) for i in itertools.combinations(all_prot_1,2)]
print(len(all_prot_2))
all_clin_1_prot_1_and_prot_2 = list(
itertools.chain(*[all_clin_1_prot_1,all_prot_2])
)
len(all_clin_1_prot_1_and_prot_2)
all_clin_2_and_clin_1_prot_1_and_prot_2 = list(
itertools.chain(*[all_clin_2,all_clin_1_prot_1,all_prot_2])
)
len(all_clin_2_and_clin_1_prot_1_and_prot_2)
all_clin_2_and_clin_1_prot_1_and_prot_2_and_clin_1_and_prot_1 = list(
itertools.chain(*[all_clin_2,all_clin_1_prot_1,all_prot_2,all_clin_1,all_prot_1])
)
print(len(all_clin_2_and_clin_1_prot_1_and_prot_2_and_clin_1_and_prot_1))
t0 = time.time()
fimps_dfs = []
perf_dfs = []
ppreds_dfs = []
perm_fimps_dfs = []
perm_perf_dfs = []
perm_ppreds_dfs = []
feature_set = {}
for i,features in enumerate(all_clin_1_and_prot_1):
if features in all_cov_df.columns:
continue
print(features)
print(i)
X_all = X_all_proteins.join(X_all_clinical)
if type(features)==np.str_:
X = X_all[[features]]
if type(features)==list:
X = X_all[features]
feature_set[str(i)] = X.columns.tolist()
params.update({'X' : X.join(all_cov_df),'models' : models.copy()})
lst = bootstrap_of_fcn(func=train_test_val_top_fold_01_within,
params=params,n_jobs=n_jobs,nboot=nboot)
perf = get_performance([lst[i][0] for i in range(len(lst))])
perf['set'] = str(i)
perf_dfs.append(perf)
fimps = model_feature_importances([lst[i][1] for i in range(len(lst))])
fimps['set'] = str(i)
fimps_dfs.append(fimps)
ppreds = patient_predictions([lst[i][2] for i in range(len(lst))])
ppreds['set'] = str(i)
ppreds_dfs.append(ppreds)
lst = bootstrap_of_fcn(func=permuted_train_test_val_top_fold_01_within,
params=params,n_jobs=n_jobs,nboot=nboot)
perm_perf = get_performance([lst[i][0] for i in range(len(lst))])
perm_perf['set'] = str(i)
perm_perf_dfs.append(perm_perf)
perm_fimps = model_feature_importances([lst[i][1] for i in range(len(lst))])
perm_fimps['set'] = str(i)
perm_fimps_dfs.append(perm_fimps)
perm_ppreds = patient_predictions([lst[i][2] for i in range(len(lst))])
perm_ppreds['set'] = str(i)
perm_ppreds_dfs.append(perm_ppreds)
perf_df = ( | pd.concat(perf_dfs) | pandas.concat |
import re
import requests
from bs4 import BeautifulSoup
import json
from collections import OrderedDict
from io import StringIO
import pandas as pd
from astropy.time import Time
from datetime import datetime,date,timedelta
from tns_api_search import search, get, format_to_json, get_file
from astropy.coordinates import SkyCoord
from astropy import units as u
url_tns_api="https://wis-tns.weizmann.ac.il/api/get"
from credentials import tns
api_key = tns.settings.API_KEY
def goodrow(class_):
return ((class_=="row-even public odd") or (class_=='row-odd public odd'))
def getTNS(reportdays=5,discoverdays=5,enddate=None,classified=1,disc_mag_min=16,disc_mag_max=21,z_min=0.015,z_max=0.08,
skip_ztf=False,num_page=100,verbose=False,otherparams={},**kwargs):
'''
returns a coma separated list with the redshift, internal name, discovery date, and discovery magnitude
of objetcs from TNS which match the search criteria
parameters:
reportdays - maximum number of days that have past since being reported
z_min - minimum redshift
z_max - maximum redshift
disc_mag_min - minimum discovery magnitude
disc_mag_max - maximum discovery magnitude
Note: I believe this is just a numerical cut, not physical, i.e. the minimum is the lowest numerical
value that will be returned
calssified - 1: is classified, 0: classification not considered
unclassified - 1: is unclassified, 0: unclassification not considered
'''
# link = f'https://wis-tns.weizmann.ac.il/search?&discovered_period_value={reportdays}&discovered_period_units=days&unclassified_at={unclassified}&classified_sne={classified}&name=&name_like=0&isTNS_AT=all&public=all&coords_unit=arcsec&redshift_min={z_min}&redshift_max={z_max}&discovery_mag_min={disc_mag_min}&discovery_mag_max={disc_mag_max}&objtype=3&sort=desc&order=discoverydate&num_page=500'
link = 'https://wis-tns.weizmann.ac.il/search'
if enddate is None:
enddate = date.today()
startdate = enddate - timedelta(discoverdays)
params = {"discovered_period_value":reportdays,
"discovered_period_units":"days",
"date_start[date]":startdate.isoformat(),
"date_end[date]":enddate.isoformat(),
"classified_sne":int(classified),
"unclassified_at":int(not(classified)),
"discovery_mag_min":disc_mag_min,
"discovery_mag_max":disc_mag_max,
"num_page":num_page
}
params.update(otherparams)
if classified:
params.update({"objtype":3,
"redshift_min":z_min,
"redshift_max":z_max,
"sort":"desc",
"order":"discoverydate"})
else:
params.update({"at_type":1,
"sort":"asc",
"order":"internal_name"})
r = requests.get(link,params=params)
if verbose:
print(r.url)
soup = BeautifulSoup(r.text, "lxml")
return_arr = []
tr = soup.find_all('tbody')
if verbose:
print("Number of tables on the webpage:",len(tr))
if len(tr)>0:
tr = tr[0]
else:
raise RuntimeError("No result is found")
cols = ['internal_name','redshift','ra','decl','hostname','host_redshift','discoverydate','discoverymag','disc_filter_name','name','ot_name']
dflist = []
if verbose:
print("Number of rows in search result: ",len(tr.find_all(class_=goodrow,recursive=False)))
for row in tr.find_all(class_=goodrow,recursive=False):
df = {}
for col in cols:
value = row.find('td',class_='cell-{}'.format(col),recursive=False)
if value is None:
df[col] = None
else:
df[col] = value.text
df['name'] = df['name'].split()[1]
if (not classified) & skip_ztf & df['internal_name'].startswith('ZTF'):
break
dflist.append(df)
df = pd.DataFrame(dflist)
df.columns = ['internal_name','redshift','ra_s','dec_s','hostname','host_redshift','discoverydate','discoverymag','disc_filter_name','tns_name','type']
c = SkyCoord(ra=df.ra_s.values, dec=df.dec_s.values, unit=(u.hourangle,u.deg))
df['meanra'] = c.ra.degree
df['meandec'] = c.dec.degree
df['oid'] = df['tns_name']
return df.sort_values('discoverydate',ascending=False).reset_index(drop=True)
def get_tns_name(internal_name):
search_obj = [("internal_name",internal_name)]
response = search(url_tns_api,search_obj)
if None not in response:
json_data = format_to_json(response.text)
reply = json.loads(json_data)['data']['reply']
if len(reply) == 0:
return
else:
return reply[0]['objname']
else:
print(response[1])
return
def get_tns_data(tns_name):
data = {}
get_obj=[("objname",tns_name), ("photometry","1"), ("spectra","1")]
response=get(url_tns_api,get_obj)
if None not in response:
# Here we just display the full json data as the response
json_data = format_to_json(response.text)
data['meta'] = format_meta(json.loads(json_data)['data']['reply'])
photometry = json.loads(json_data)['data']['reply']['photometry']
spectra = json.loads(json_data)['data']['reply']['spectra']
data['photometry'] = format_photometry(photometry)
data['spectra'] = format_spectra(spectra)
else:
print (response[1])
data = None
return data
def format_meta(reply):
cols = ['internal_name','redshift','radeg','decdeg','hostname','host_redshift','discoverydate','discoverymag','discmagfilter','name']
df = {k: reply[k] for k in cols}
return pd.DataFrame([ | pd.DataFrame(df) | pandas.DataFrame |
# -*- coding: utf-8 -*-
"""
Created on Wed Nov 28 17:05:36 2018
@author: kutay.erkan
"""
"""
References:
https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html
https://scikit-learn.org/stable/auto_examples/decomposition/plot_pca_vs_lda.html
https://seaborn.pydata.org/generated/seaborn.lmplot.html
https://cmdlinetips.com/2018/03/pca-example-in-python-with-scikit-learn/
"""
import os
os.getcwd()
# %% Import libraries
import numpy as np
import pandas as pd
import seaborn as sns
from sklearn.model_selection import train_test_split
# from sklearn.feature_selection import SelectFromModel
from sklearn.neighbors import KNeighborsClassifier
from sklearn.decomposition import PCA
from sklearn.metrics import accuracy_score, recall_score, precision_score, confusion_matrix
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "last_expr" # "all" to print multiple statements
# %% Read data
df = pd.read_table("dataset.txt", sep=",", header=None)
# Name features
df.columns = ["word_freq_make", "word_freq_address", "word_freq_all",
"word_freq_3d", "word_freq_our", "word_freq_over",
"word_freq_remove", "word_freq_internet", "word_freq_order",
"word_freq_mail", "word_freq_receive", "word_freq_will",
"word_freq_people", "word_freq_report", "word_freq_addresses",
"word_freq_free", "word_freq_business", "word_freq_email",
"word_freq_you", "word_freq_credit", "word_freq_your",
"word_freq_font", "word_freq_000", "word_freq_money",
"word_freq_hp", "word_freq_hpl", "word_freq_george",
"word_freq_650", "word_freq_lab", "word_freq_labs",
"word_freq_telnet", "word_freq_857", "word_freq_data",
"word_freq_415", "word_freq_85", "word_freq_technology",
"word_freq_1999", "word_freq_parts", "word_freq_pm",
"word_freq_direct", "word_freq_cs", "word_freq_meeting",
"word_freq_original", "word_freq_project", "word_freq_re",
"word_freq_edu", "word_freq_table", "word_freq_conference",
"char_freq_;", "char_freq_(", "char_freq_[", "char_freq_!",
"char_freq_$", "char_freq_#", "capital_run_length_average",
"capital_run_length_longest", "capital_run_length_total",
"is_spam"
]
# %% Explore data
print (df.head(),"\n")
print ("Number of all instances: {}".format(len(df)))
print ("Number of spam instances: {}\n".format(df.is_spam.sum()))
print ("Features:\n {}".format(list(df)))
# %% Name the label and features
label = df.is_spam
features = df.drop(columns=['is_spam'])
# %% Train/Test Split
features_train, features_test, label_train, label_test = train_test_split(features,label, test_size=0.5, random_state = 22, stratify=df.is_spam)
print ("Number of spam instances in training set: {}".format(label_train.sum()))
print ("Number of spam instances in test set: {}".format(label_test.sum()))
# %% Directly run kNN on training and test data without PCA or feature reduction
clf = KNeighborsClassifier(n_neighbors=5, metric='euclidean')
clf.fit(features_train, label_train)
label_train_pred = clf.predict(features_train)
label_test_pred = clf.predict(features_test)
print ("\nPerformance on Training Data without Feature Reduction:")
print ("Accuracy: {}".format(accuracy_score(label_train, label_train_pred)))
print ("Recall: {}".format(recall_score(label_train, label_train_pred)))
print ("Precision: {}".format(precision_score(label_train, label_train_pred)))
print ("Confusion Matrix: \n{}\n".format(confusion_matrix(label_train, label_train_pred)))
print ("Performance on Test Data without Feature Reduction:")
print ("Accuracy: {}".format(accuracy_score(label_test, label_test_pred)))
print ("Recall: {}".format(recall_score(label_test, label_test_pred)))
print ("Precision: {}".format(precision_score(label_test, label_test_pred)))
print ("Confusion Matrix: \n{}\n".format(confusion_matrix(label_test, label_test_pred)))
# %% See accuracy score and cumulative explained variance for different m using training data
selected_n = 0
pca_accuracy = 0
for i in range(1, 58):
pca = PCA(n_components=i)
pca.fit(features_train)
pc = pca.fit_transform(features_train)
clf.fit(pc,label_train)
label_train_pred = clf.predict(pc)
if accuracy_score(label_train, label_train_pred) > pca_accuracy:
pca_accuracy = accuracy_score(label_train, label_train_pred)
selected_n = i
print ("Accuracy for {} features: {}".format(i,accuracy_score(label_train, label_train_pred)))
print ("Recall for {} features: {}".format(i,recall_score(label_train, label_train_pred)))
print ("Precision for {} features: {}".format(i,precision_score(label_train, label_train_pred)))
print ("Cum. explained variance ratio for {} features: {}".format(i,pca.explained_variance_ratio_.cumsum()[-1]))
print ("\nSelected n_components with highest accuracy score: {}".format(selected_n))
print ("Accuracy score for {} components: {}".format(selected_n, pca_accuracy))
# %% Plot for m=2
pca = PCA(n_components=2)
pc = pca.fit_transform(features_train)
pc_df = pd.DataFrame(data = pc, columns = ['PC1', 'PC2'])
label_train=label_train.reset_index(drop=True)
pc_df = pd.concat([pc_df,label_train],axis=1)
print ("\nPlot for 2 principal components PC1, PC2:")
sns.lmplot(x="PC1", y="PC2", data=pc_df, hue='is_spam',
fit_reg=False, scatter_kws={"alpha": 0.2});
# %% See performance metrics for chosen m using training and test data
print('n_components=41 and 42 have the same accuracy. Which one is selected DOES change on runtime.')
print ("\nSelected n_components: {}".format(selected_n))
pca = PCA(n_components=selected_n)
pca.fit(features_train)
pc = pca.fit_transform(features_train)
clf.fit(pc,label_train)
label_train_pred = clf.predict(pc)
print ("\nPerformance on Training Data with Feature Reduction Using PCA:")
print ("Accuracy: {}".format(accuracy_score(label_train, label_train_pred)))
print ("Recall: {}".format(recall_score(label_train, label_train_pred)))
print ("Precision: {}".format(precision_score(label_train, label_train_pred)))
print ("Confusion Matrix: \n{}\n".format(confusion_matrix(label_train, label_train_pred)))
pca = PCA(n_components=42)
pca.fit(features_test)
pc = pca.fit_transform(features_test)
clf.fit(pc,label_test)
label_test_pred = clf.predict(pc)
print ("Performance on Test Data with Feature Reduction Using PCA:")
print ("Accuracy: {}".format(accuracy_score(label_test, label_test_pred)))
print ("Recall: {}".format(recall_score(label_test, label_test_pred)))
print ("Precision: {}".format(precision_score(label_test, label_test_pred)))
print ("Confusion Matrix: \n{}\n".format(confusion_matrix(label_test, label_test_pred)))
# %% Finding feature accuracies
clf = KNeighborsClassifier(n_neighbors=5, metric='euclidean')
knn_accuracy = 0
accuracy_list = | pd.DataFrame(columns=["feature","accuracy"]) | pandas.DataFrame |
# coding: utf-8
# # CareerCon 2019 - Help Navigate Robots
# ## Robots are smart… by design !!
#
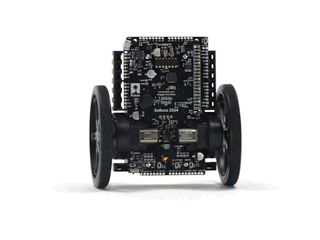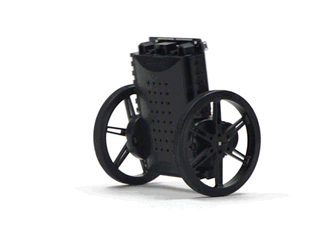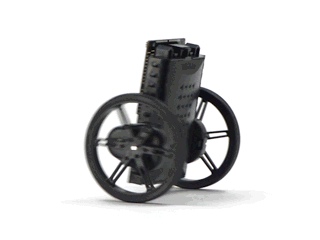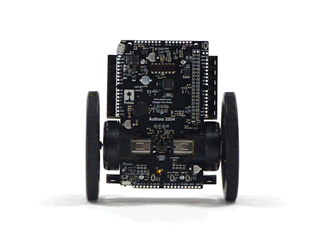
# 
#
# ---
#
# Robots are smart… by design. To fully understand and properly navigate a task, however, they need input about their environment.
#
# In this competition, you’ll help robots recognize the floor surface they’re standing on using data collected from Inertial Measurement Units (IMU sensors).
#
# We’ve collected IMU sensor data while driving a small mobile robot over different floor surfaces on the university premises. The task is to predict which one of the nine floor types (carpet, tiles, concrete) the robot is on using sensor data such as acceleration and velocity. Succeed and you'll help improve the navigation of robots without assistance across many different surfaces, so they won’t fall down on the job.
#
# ### Its a golden chance to help humanity, by helping Robots !
#
# <br>
# <img src="https://media2.giphy.com/media/EizPK3InQbrNK/giphy.gif" border="1" width="400" height="300">
# <br>
# # DATA
# **X_[train/test].csv** - the input data, covering 10 sensor channels and 128 measurements per time series plus three ID columns:
#
# - ```row_id```: The ID for this row.
#
# - ```series_id: ID``` number for the measurement series. Foreign key to y_train/sample_submission.
#
# - ```measurement_number```: Measurement number within the series.
#
# The orientation channels encode the current angles how the robot is oriented as a quaternion (see Wikipedia). Angular velocity describes the angle and speed of motion, and linear acceleration components describe how the speed is changing at different times. The 10 sensor channels are:
#
# ```
# orientation_X
#
# orientation_Y
#
# orientation_Z
#
# orientation_W
#
# angular_velocity_X
#
# angular_velocity_Y
#
# angular_velocity_Z
#
# linear_acceleration_X
#
# linear_acceleration_Y
#
# linear_acceleration_Z
# ```
#
# **y_train.csv** - the surfaces for training set.
#
# - ```series_id```: ID number for the measurement series.
#
# - ```group_id```: ID number for all of the measurements taken in a recording session. Provided for the training set only, to enable more cross validation strategies.
#
# - ```surface```: the target for this competition.
#
# **sample_submission.csv** - a sample submission file in the correct format.
# ### Load packages
# In[1]:
import numpy as np
import pandas as pd
import os
from time import time
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from scipy.stats import norm
from sklearn.preprocessing import StandardScaler
from matplotlib import rcParams
get_ipython().run_line_magic('matplotlib', 'inline')
le = preprocessing.LabelEncoder()
from numba import jit
import itertools
from seaborn import countplot,lineplot, barplot
from numba import jit
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
from sklearn import preprocessing
from scipy.stats import randint as sp_randint
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import KFold
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import LeaveOneGroupOut
from sklearn.model_selection import GroupKFold
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import RandomizedSearchCV
import matplotlib.style as style
style.use('ggplot')
import warnings
warnings.filterwarnings('ignore')
import gc
gc.enable()
get_ipython().system('ls ../input/')
get_ipython().system('ls ../input/robots-best-submission')
print ("Ready !")
# ### Load data
# In[2]:
data = pd.read_csv('../input/career-con-2019/X_train.csv')
tr = pd.read_csv('../input/career-con-2019/X_train.csv')
sub = pd.read_csv('../input/career-con-2019/sample_submission.csv')
test = pd.read_csv('../input/career-con-2019/X_test.csv')
target = pd.read_csv('../input/career-con-2019/y_train.csv')
print ("Data is ready !!")
# # Data exploration
# In[3]:
data.head()
# In[4]:
test.head()
# In[5]:
target.head()
# In[6]:
len(data.measurement_number.value_counts())
# Each series has 128 measurements.
#
# **1 serie = 128 measurements**.
#
# For example, serie with series_id=0 has a surface = *fin_concrete* and 128 measurements.
# ### describe (basic stats)
# In[7]:
data.describe()
# In[8]:
test.describe()
# In[9]:
target.describe()
# ### There is missing data in test and train data
# In[10]:
totalt = data.isnull().sum().sort_values(ascending=False)
percent = (data.isnull().sum()/data.isnull().count()).sort_values(ascending=False)
missing_data = pd.concat([totalt, percent], axis=1, keys=['Total', 'Percent'])
print ("Missing Data at Training")
missing_data.tail()
# In[11]:
totalt = test.isnull().sum().sort_values(ascending=False)
percent = (test.isnull().sum()/data.isnull().count()).sort_values(ascending=False)
missing_data = pd.concat([totalt, percent], axis=1, keys=['Total', 'Percent'])
print ("Missing Data at Test")
missing_data.tail()
# In[12]:
print ("Test has ", (test.shape[0]-data.shape[0])/128, "series more than Train (later I will prove it) = 768 registers")
dif = test.shape[0]-data.shape[0]
print ("Let's check this extra 6 series")
test.tail(768).describe()
# If we look at the features: orientation, angular velocity and linear acceleration, we can see big differences between **max** and **min** from entire test vs 6 extra test's series (see **linear_acceleration_Z**).
#
# Obviously we are comparing 3810 series vs 6 series so this is not a big deal.
# ### goup_id will be important !!
# In[13]:
target.groupby('group_id').surface.nunique().max()
# In[14]:
target['group_id'].nunique()
# **73 groups**
# **Each group_id is a unique recording session and has only one surface type **
# In[15]:
sns.set(style='darkgrid')
sns.countplot(y = 'surface',
data = target,
order = target['surface'].value_counts().index)
plt.show()
# ### Target feature - surface and group_id distribution
# Let's show now the distribution of target feature - surface and group_id.
# by @gpreda.
# In[16]:
fig, ax = plt.subplots(1,1,figsize=(26,8))
tmp = pd.DataFrame(target.groupby(['group_id', 'surface'])['series_id'].count().reset_index())
m = tmp.pivot(index='surface', columns='group_id', values='series_id')
s = sns.heatmap(m, linewidths=.1, linecolor='black', annot=True, cmap="YlGnBu")
s.set_title('Number of surface category per group_id', size=16)
plt.show()
# We need to classify on which surface our robot is standing.
#
# Multi-class Multi-output
#
# 9 classes (suface)
# In[17]:
plt.figure(figsize=(23,5))
sns.set(style="darkgrid")
countplot(x="group_id", data=target, order = target['group_id'].value_counts().index)
plt.show()
# **So, we have 3810 train series, and 3816 test series.
# Let's engineer some features!**
#
# ## Example: Series 1
#
# Let's have a look at the values of features in a single time-series, for example series 1 ```series_id=0```
#
# Click to see all measurements of the **first series**
# In[18]:
serie1 = tr.head(128)
serie1.head()
# In[19]:
serie1.describe()
# In[20]:
plt.figure(figsize=(26, 16))
for i, col in enumerate(serie1.columns[3:]):
plt.subplot(3, 4, i + 1)
plt.plot(serie1[col])
plt.title(col)
# In this example, we can see a quite interesting performance:
# 1. Orientation X increases
# 2. Orientation Y decreases
# 3. We don't see any kind of pattern except for linear_acceleration_Y
#
# And we know that in this series, the robot moved throuh "fine_concrete".
# In[21]:
target.head(1)
# In[22]:
del serie1
gc.collect()
# ## Visualizing Series
#
# Before, I showed you as an example the series 1.
#
# **This code allows you to visualize any series.**
#
# From: *Code Snippet For Visualizing Series Id by @shaz13*
# In[23]:
series_dict = {}
for series in (data['series_id'].unique()):
series_dict[series] = data[data['series_id'] == series]
# In[24]:
def plotSeries(series_id):
style.use('ggplot')
plt.figure(figsize=(28, 16))
print(target[target['series_id'] == series_id]['surface'].values[0].title())
for i, col in enumerate(series_dict[series_id].columns[3:]):
if col.startswith("o"):
color = 'red'
elif col.startswith("a"):
color = 'green'
else:
color = 'blue'
if i >= 7:
i+=1
plt.subplot(3, 4, i + 1)
plt.plot(series_dict[series_id][col], color=color, linewidth=3)
plt.title(col)
# **Now, Let's see code for series 15 ( is an example, try what you want)**
# In[25]:
id_series = 15
plotSeries(id_series)
# In[26]:
del series_dict
gc.collect()
# <br>
# ### Correlations (Part I)
# In[27]:
f,ax = plt.subplots(figsize=(8, 8))
sns.heatmap(tr.iloc[:,3:].corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax)
# **Correlations test (click "code")**
# In[28]:
f,ax = plt.subplots(figsize=(8, 8))
sns.heatmap(test.iloc[:,3:].corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax)
# Well, this is immportant, there is a **strong correlation** between:
# - angular_velocity_Z and angular_velocity_Y
# - orientation_X and orientation_Y
# - orientation_Y and orientation_Z
#
# Moreover, test has different correlations than training, for example:
#
# - angular_velocity_Z and orientation_X: -0.1(training) and 0.1(test). Anyway, is too small in both cases, it should not be a problem.
# ## Fourier Analysis
#
# My hope was, that different surface types yield (visible) differences in the frequency spectrum of the sensor measurements.
#
# Machine learning techniques might learn frequency filters on their own, but why don't give the machine a little head start? So I computed the the cyclic FFT for the angular velocity and linear acceleration sensors and plotted mean and standard deviation of the absolute values of the frequency components per training surface category (leaving out the frequency 0 (i.e. constants like sensor bias, earth gravity, ...).
#
# The sensors show some different frequency characterists (see plots below), but unfortunately the surface categories have all similar (to the human eye) shapes, varying mostly in total power, and the standard deviations are high (compared to differences in the means). So there are no nice strong characteristic peaks for surface types. But that does not mean, that there is nothing detectable by more sophisticated statistical methods.
#
# This article http://www.kaggle.com/christoffer/establishing-sampling-frequency makes a convincing case, that the sampling frequency is around 400Hz, so according to that you would see the frequency range to 3-200 Hz in the diagrams (and aliased higher frequencies).
#
# by [@trohwer64](https://www.kaggle.com/trohwer64)
# In[29]:
get_ipython().system('ls ../input')
# In[30]:
train_x = pd.read_csv('../input/career-con-2019/X_train.csv')
train_y = pd.read_csv('../input/career-con-2019/y_train.csv')
# In[31]:
import math
def prepare_data(t):
def f(d):
d=d.sort_values(by=['measurement_number'])
return pd.DataFrame({
'lx':[ d['linear_acceleration_X'].values ],
'ly':[ d['linear_acceleration_Y'].values ],
'lz':[ d['linear_acceleration_Z'].values ],
'ax':[ d['angular_velocity_X'].values ],
'ay':[ d['angular_velocity_Y'].values ],
'az':[ d['angular_velocity_Z'].values ],
})
t= t.groupby('series_id').apply(f)
def mfft(x):
return [ x/math.sqrt(128.0) for x in np.absolute(np.fft.fft(x)) ][1:65]
t['lx_f']=[ mfft(x) for x in t['lx'].values ]
t['ly_f']=[ mfft(x) for x in t['ly'].values ]
t['lz_f']=[ mfft(x) for x in t['lz'].values ]
t['ax_f']=[ mfft(x) for x in t['ax'].values ]
t['ay_f']=[ mfft(x) for x in t['ay'].values ]
t['az_f']=[ mfft(x) for x in t['az'].values ]
return t
# In[32]:
t=prepare_data(train_x)
t=pd.merge(t,train_y[['series_id','surface','group_id']],on='series_id')
t=t.rename(columns={"surface": "y"})
# In[33]:
def aggf(d, feature):
va= np.array(d[feature].tolist())
mean= sum(va)/va.shape[0]
var= sum([ (va[i,:]-mean)**2 for i in range(va.shape[0]) ])/va.shape[0]
dev= [ math.sqrt(x) for x in var ]
return pd.DataFrame({
'mean': [ mean ],
'dev' : [ dev ],
})
display={
'hard_tiles_large_space':'r-.',
'concrete':'g-.',
'tiled':'b-.',
'fine_concrete':'r-',
'wood':'g-',
'carpet':'b-',
'soft_pvc':'y-',
'hard_tiles':'r--',
'soft_tiles':'g--',
}
# In[34]:
import matplotlib.pyplot as plt
plt.figure(figsize=(14, 8*7))
#plt.margins(x=0.0, y=0.0)
#plt.tight_layout()
# plt.figure()
features=['lx_f','ly_f','lz_f','ax_f','ay_f','az_f']
count=0
for feature in features:
stat= t.groupby('y').apply(aggf,feature)
stat.index= stat.index.droplevel(-1)
b=[*range(len(stat.at['carpet','mean']))]
count+=1
plt.subplot(len(features)+1,1,count)
for i,(k,v) in enumerate(display.items()):
plt.plot(b, stat.at[k,'mean'], v, label=k)
# plt.errorbar(b, stat.at[k,'mean'], yerr=stat.at[k,'dev'], fmt=v)
leg = plt.legend(loc='best', ncol=3, mode="expand", shadow=True, fancybox=True)
plt.title("sensor: " + feature)
plt.xlabel("frequency component")
plt.ylabel("amplitude")
count+=1
plt.subplot(len(features)+1,1,count)
k='concrete'
v=display[k]
feature='lz_f'
stat= t.groupby('y').apply(aggf,feature)
stat.index= stat.index.droplevel(-1)
b=[*range(len(stat.at['carpet','mean']))]
plt.errorbar(b, stat.at[k,'mean'], yerr=stat.at[k,'dev'], fmt=v)
plt.title("sample for error bars (lz_f, surface concrete)")
plt.xlabel("frequency component")
plt.ylabel("amplitude")
plt.show()
# In[35]:
del train_x, train_y
gc.collect()
# ## Is it an Humanoid Robot instead of a car?
#
# 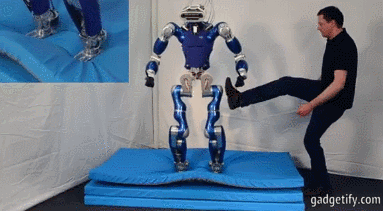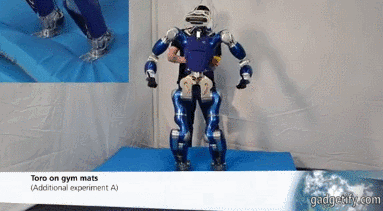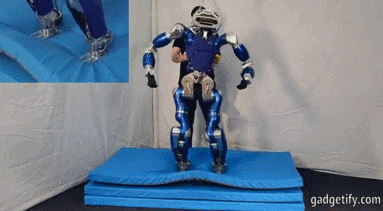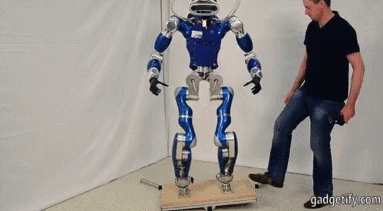
#
# **Acceleration**
# - X (mean at 0)
# - Y axis is centered at a value wich shows us the movement (straight ).
# - Z axis is centered at 10 (+- 9.8) wich is the gravity !! , you can see how the robot bounds.
#
# Angular velocity (X,Y,Z) has mean (0,0,0) so there is no lineal movement on those axis (measured with an encoder or potentiometer)
#
# **Fourier**
#
# We can see: with a frequency 3 Hz we can see an acceleration, I think that acceleration represents one step.
# Maybe ee can suppose that every step is caused by many different movements, that's why there are different accelerations at different frequencies.
#
# Angular velocity represents spins.
# Every time the engine/servo spins, the robot does an step - relation between acc y vel.
# ---
#
# # Feature Engineering
# In[36]:
def plot_feature_distribution(df1, df2, label1, label2, features,a=2,b=5):
i = 0
sns.set_style('whitegrid')
plt.figure()
fig, ax = plt.subplots(a,b,figsize=(17,9))
for feature in features:
i += 1
plt.subplot(a,b,i)
sns.kdeplot(df1[feature], bw=0.5,label=label1)
sns.kdeplot(df2[feature], bw=0.5,label=label2)
plt.xlabel(feature, fontsize=9)
locs, labels = plt.xticks()
plt.tick_params(axis='x', which='major', labelsize=8)
plt.tick_params(axis='y', which='major', labelsize=8)
plt.show();
# In[37]:
features = data.columns.values[3:]
plot_feature_distribution(data, test, 'train', 'test', features)
# Godd news, our basic features have the **same distribution (Normal) on test and training**. There are some differences between *orientation_X* , *orientation_Y* and *linear_acceleration_Y*.
#
# I willl try **StandardScaler** to fix this, and remember: orientation , angular velocity and linear acceleration are measured with different units, scaling might be a good choice.
# In[38]:
def plot_feature_class_distribution(classes,tt, features,a=5,b=2):
i = 0
sns.set_style('whitegrid')
plt.figure()
fig, ax = plt.subplots(a,b,figsize=(16,24))
for feature in features:
i += 1
plt.subplot(a,b,i)
for clas in classes:
ttc = tt[tt['surface']==clas]
sns.kdeplot(ttc[feature], bw=0.5,label=clas)
plt.xlabel(feature, fontsize=9)
locs, labels = plt.xticks()
plt.tick_params(axis='x', which='major', labelsize=8)
plt.tick_params(axis='y', which='major', labelsize=8)
plt.show();
# In[39]:
classes = (target['surface'].value_counts()).index
aux = data.merge(target, on='series_id', how='inner')
plot_feature_class_distribution(classes, aux, features)
# **Normal distribution**
#
# There are obviously differences between *surfaces* and that's good, we will focus on that in order to classify them better.
#
# Knowing this differences and that variables follow a normal distribution (in most of the cases) we need to add new features like: ```mean, std, median, range ...``` (for each variable).
#
# However, I will try to fix *orientation_X* and *orientation_Y* as I explained before, scaling and normalizing data.
#
# ---
#
# ### Now with a new scale (more more precision)
# In[40]:
plt.figure(figsize=(26, 16))
for i,col in enumerate(aux.columns[3:13]):
ax = plt.subplot(3,4,i+1)
ax = plt.title(col)
for surface in classes:
surface_feature = aux[aux['surface'] == surface]
sns.kdeplot(surface_feature[col], label = surface)
# ### Histogram for main features
# In[41]:
plt.figure(figsize=(26, 16))
for i, col in enumerate(data.columns[3:]):
ax = plt.subplot(3, 4, i + 1)
sns.distplot(data[col], bins=100, label='train')
sns.distplot(test[col], bins=100, label='test')
ax.legend()
# ## Step 0 : quaternions
# Orientation - quaternion coordinates
# You could notice that there are 4 coordinates: X, Y, Z, W.
#
# Usually we have X, Y, Z - Euler Angles. But Euler Angles are limited by a phenomenon called "gimbal lock," which prevents them from measuring orientation when the pitch angle approaches +/- 90 degrees. Quaternions provide an alternative measurement technique that does not suffer from gimbal lock. Quaternions are less intuitive than Euler Angles and the math can be a little more complicated.
#
# Here are some articles about it:
#
# http://www.chrobotics.com/library/understanding-quaternions
#
# http://www.tobynorris.com/work/prog/csharp/quatview/help/orientations_and_quaternions.htm
#
# Basically 3D coordinates are converted to 4D vectors.
# In[42]:
# https://stackoverflow.com/questions/53033620/how-to-convert-euler-angles-to-quaternions-and-get-the-same-euler-angles-back-fr?rq=1
def quaternion_to_euler(x, y, z, w):
import math
t0 = +2.0 * (w * x + y * z)
t1 = +1.0 - 2.0 * (x * x + y * y)
X = math.atan2(t0, t1)
t2 = +2.0 * (w * y - z * x)
t2 = +1.0 if t2 > +1.0 else t2
t2 = -1.0 if t2 < -1.0 else t2
Y = math.asin(t2)
t3 = +2.0 * (w * z + x * y)
t4 = +1.0 - 2.0 * (y * y + z * z)
Z = math.atan2(t3, t4)
return X, Y, Z
# In[43]:
def fe_step0 (actual):
# https://www.mathworks.com/help/aeroblks/quaternionnorm.html
# https://www.mathworks.com/help/aeroblks/quaternionmodulus.html
# https://www.mathworks.com/help/aeroblks/quaternionnormalize.html
# Spoiler: you don't need this ;)
actual['norm_quat'] = (actual['orientation_X']**2 + actual['orientation_Y']**2 + actual['orientation_Z']**2 + actual['orientation_W']**2)
actual['mod_quat'] = (actual['norm_quat'])**0.5
actual['norm_X'] = actual['orientation_X'] / actual['mod_quat']
actual['norm_Y'] = actual['orientation_Y'] / actual['mod_quat']
actual['norm_Z'] = actual['orientation_Z'] / actual['mod_quat']
actual['norm_W'] = actual['orientation_W'] / actual['mod_quat']
return actual
#
# > *Are there any reasons to not automatically normalize a quaternion? And if there are, what quaternion operations do result in non-normalized quaternions?*
#
# Any operation that produces a quaternion will need to be normalized because floating-point precession errors will cause it to not be unit length.
# I would advise against standard routines performing normalization automatically for performance reasons.
# Any competent programmer should be aware of the precision issues and be able to normalize the quantities when necessary - and it is not always necessary to have a unit length quaternion.
# The same is true for vector operations.
#
# source: https://stackoverflow.com/questions/11667783/quaternion-and-normalization
# In[44]:
data = fe_step0(data)
test = fe_step0(test)
print(data.shape)
data.head()
# In[45]:
fig, (ax1, ax2, ax3, ax4) = plt.subplots(ncols=4, figsize=(18, 5))
ax1.set_title('quaternion X')
sns.kdeplot(data['norm_X'], ax=ax1, label="train")
sns.kdeplot(test['norm_X'], ax=ax1, label="test")
ax2.set_title('quaternion Y')
sns.kdeplot(data['norm_Y'], ax=ax2, label="train")
sns.kdeplot(test['norm_Y'], ax=ax2, label="test")
ax3.set_title('quaternion Z')
sns.kdeplot(data['norm_Z'], ax=ax3, label="train")
sns.kdeplot(test['norm_Z'], ax=ax3, label="test")
ax4.set_title('quaternion W')
sns.kdeplot(data['norm_W'], ax=ax4, label="train")
sns.kdeplot(test['norm_W'], ax=ax4, label="test")
plt.show()
# ## Step 1: (x, y, z, w) -> (x,y,z) quaternions to euler angles
# In[46]:
def fe_step1 (actual):
"""Quaternions to Euler Angles"""
x, y, z, w = actual['norm_X'].tolist(), actual['norm_Y'].tolist(), actual['norm_Z'].tolist(), actual['norm_W'].tolist()
nx, ny, nz = [], [], []
for i in range(len(x)):
xx, yy, zz = quaternion_to_euler(x[i], y[i], z[i], w[i])
nx.append(xx)
ny.append(yy)
nz.append(zz)
actual['euler_x'] = nx
actual['euler_y'] = ny
actual['euler_z'] = nz
return actual
# In[47]:
data = fe_step1(data)
test = fe_step1(test)
print (data.shape)
data.head()
# 
# In[48]:
fig, (ax1, ax2, ax3) = plt.subplots(ncols=3, figsize=(15, 5))
ax1.set_title('Roll')
sns.kdeplot(data['euler_x'], ax=ax1, label="train")
sns.kdeplot(test['euler_x'], ax=ax1, label="test")
ax2.set_title('Pitch')
sns.kdeplot(data['euler_y'], ax=ax2, label="train")
sns.kdeplot(test['euler_y'], ax=ax2, label="test")
ax3.set_title('Yaw')
sns.kdeplot(data['euler_z'], ax=ax3, label="train")
sns.kdeplot(test['euler_z'], ax=ax3, label="test")
plt.show()
# **Euler angles** are really important, and we have a problem with Z.
#
# ### Why Orientation_Z (euler angle Z) is so important?
#
# We have a robot moving around, imagine a robot moving straight through different surfaces (each with different features), for example concrete and hard tile floor. Our robot can can **bounce** or **balance** itself a little bit on if the surface is not flat and smooth, that's why we need to work with quaternions and take care of orientation_Z.
#
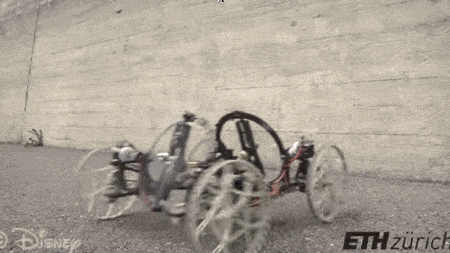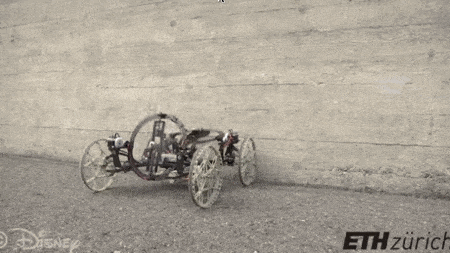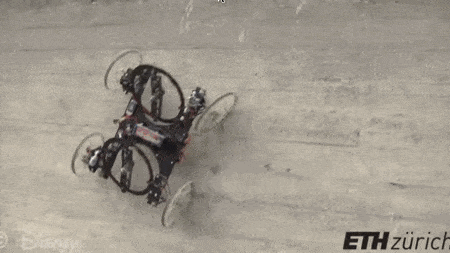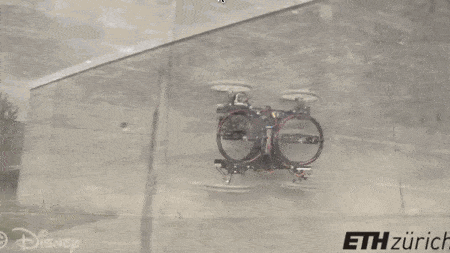
# 
# In[49]:
data.head()
# ## Step 2: + Basic features
# In[50]:
def feat_eng(data):
df = pd.DataFrame()
data['totl_anglr_vel'] = (data['angular_velocity_X']**2 + data['angular_velocity_Y']**2 + data['angular_velocity_Z']**2)** 0.5
data['totl_linr_acc'] = (data['linear_acceleration_X']**2 + data['linear_acceleration_Y']**2 + data['linear_acceleration_Z']**2)**0.5
data['totl_xyz'] = (data['orientation_X']**2 + data['orientation_Y']**2 + data['orientation_Z']**2)**0.5
data['acc_vs_vel'] = data['totl_linr_acc'] / data['totl_anglr_vel']
def mean_change_of_abs_change(x):
return np.mean(np.diff(np.abs(np.diff(x))))
for col in data.columns:
if col in ['row_id','series_id','measurement_number']:
continue
df[col + '_mean'] = data.groupby(['series_id'])[col].mean()
df[col + '_median'] = data.groupby(['series_id'])[col].median()
df[col + '_max'] = data.groupby(['series_id'])[col].max()
df[col + '_min'] = data.groupby(['series_id'])[col].min()
df[col + '_std'] = data.groupby(['series_id'])[col].std()
df[col + '_range'] = df[col + '_max'] - df[col + '_min']
df[col + '_maxtoMin'] = df[col + '_max'] / df[col + '_min']
df[col + '_mean_abs_chg'] = data.groupby(['series_id'])[col].apply(lambda x: np.mean(np.abs(np.diff(x))))
df[col + '_mean_change_of_abs_change'] = data.groupby('series_id')[col].apply(mean_change_of_abs_change)
df[col + '_abs_max'] = data.groupby(['series_id'])[col].apply(lambda x: np.max(np.abs(x)))
df[col + '_abs_min'] = data.groupby(['series_id'])[col].apply(lambda x: np.min(np.abs(x)))
df[col + '_abs_avg'] = (df[col + '_abs_min'] + df[col + '_abs_max'])/2
return df
# In[51]:
get_ipython().run_cell_magic('time', '', 'data = feat_eng(data)\ntest = feat_eng(test)\nprint ("New features: ",data.shape)')
# In[52]:
data.head()
# ## New advanced features
# **Useful functions**
# In[53]:
from scipy.stats import kurtosis
from scipy.stats import skew
def _kurtosis(x):
return kurtosis(x)
def CPT5(x):
den = len(x)*np.exp(np.std(x))
return sum(np.exp(x))/den
def skewness(x):
return skew(x)
def SSC(x):
x = np.array(x)
x = np.append(x[-1], x)
x = np.append(x,x[1])
xn = x[1:len(x)-1]
xn_i2 = x[2:len(x)] # xn+1
xn_i1 = x[0:len(x)-2] # xn-1
ans = np.heaviside((xn-xn_i1)*(xn-xn_i2),0)
return sum(ans[1:])
def wave_length(x):
x = np.array(x)
x = np.append(x[-1], x)
x = np.append(x,x[1])
xn = x[1:len(x)-1]
xn_i2 = x[2:len(x)] # xn+1
return sum(abs(xn_i2-xn))
def norm_entropy(x):
tresh = 3
return sum(np.power(abs(x),tresh))
def SRAV(x):
SRA = sum(np.sqrt(abs(x)))
return np.power(SRA/len(x),2)
def mean_abs(x):
return sum(abs(x))/len(x)
def zero_crossing(x):
x = np.array(x)
x = np.append(x[-1], x)
x = np.append(x,x[1])
xn = x[1:len(x)-1]
xn_i2 = x[2:len(x)] # xn+1
return sum(np.heaviside(-xn*xn_i2,0))
# This advanced features based on robust statistics.
# In[54]:
def fe_advanced_stats(data):
df = | pd.DataFrame() | pandas.DataFrame |
import viola
import pandas as pd
from io import StringIO
import sys, os
HERE = os.path.abspath(os.path.dirname(__file__))
data_expected = """test1 0 small_del
test2 0 small_del
test3 0 large_del
test4 0 large_del
test5 0 large_del
test6 0 small_dup
test7 0 small_inv
test8 0 others
test9 0 small_inv
viola_breakpoint:0 0 tra
viola_breakpoint:1 0 tra
"""
DEFINITIONS = """name 'small_del'
0 SVLEN > -100
1 SVTYPE == DEL
logic 0 & 1
name 'large_del'
0 SVTYPE == DEL
logic 0
name 'small_dup'
0 SVLEN < 100
1 SVTYPE == DUP
logic 0 & 1
name 'large_dup'
0 SVTYPE == DUP
logic 0
name 'small_inv'
0 SVLEN < 100
1 SVTYPE == INV
logic 0 & 1
name 'tra'
0 SVTYPE == TRA
logic 0
"""
def small_del(x):
return x.filter(['svlen > -100', 'svtype == DEL']).ids
def large_del(x):
return x.filter(['svtype == DEL']).ids
def small_dup(x):
return x.filter(['svlen < 100', 'svtype == DUP']).ids
def large_dup(x):
return x.filter(['svtype == DUP']).ids
def small_inv(x):
return x.filter(['svlen < 100', 'svtype == INV']).ids
def tra(x):
return x.filter('svtype == TRA').ids
def test_classify_manual_svtype():
vcf = viola.read_vcf(os.path.join(HERE, 'data/manta1.vcf'))
vcf = vcf.breakend2breakpoint()
ls_conditions = [small_del, large_del, small_dup, large_dup, small_inv, tra]
ls_names = ['small_del', 'large_del', 'small_dup', 'large_dup', 'small_inv', 'tra']
result = vcf.classify_manual_svtype(ls_conditions=ls_conditions, ls_names=ls_names)
manual_sv_type = vcf.manual_sv_type
manual_sv_type.set_index('id', inplace=True)
manual_sv_type_expected = pd.read_csv(StringIO(data_expected), sep='\t', names=('id', 'value_idx', 'manual_sv_type'))
manual_sv_type_expected.set_index('id', inplace=True)
pd.testing.assert_frame_equal(manual_sv_type, manual_sv_type_expected, check_like=True)
result_expected = | pd.Series([2, 3, 1, 0, 2, 2, 1]) | pandas.Series |
#!/usr/bin/env python3
# various functions and mixins for downstream genomic and epigenomic anlyses
import os
import glob
import re
import random
from datetime import datetime
import time
from pybedtools import BedTool
import pandas as pd
import numpy as np
from tqdm import tqdm_notebook, tqdm
# Get Current Git Commit Hash for version
path = [x.replace(' ', r'\ ') for x in os.popen('echo $PYTHONPATH').read().split(':') if 'dkfunctions' in x.split('/')]
if len(path) > 0:
version = os.popen(f'cd {path[0]}; git rev-parse HEAD').read()[:-1]
__version__ = f'v0.1, Git SHA1: {version}'
else:
__version__ = f'v0.1, {datetime.now():%Y-%m-%d}'
def val_folder(folder):
folder = folder if folder.endswith('/') else f'{folder}/'
folder = f'{os.getcwd()}/' if folder == '/' else folder
os.makedirs(folder, exist_ok=True)
return folder
def image_display(file):
from IPython.display import Image, display
display(Image(file))
def rplot(plot_func, filename, filetype, *args, **kwargs):
from rpy2.robjects.packages import importr
grdevices = importr('grDevices')
filetype = filetype.lower()
plot_types = {'png': grdevices.png,
'svg': grdevices.svg,
'pdf': grdevices.pdf
}
plot_types[filetype](f'{filename}.{filetype}')
return_object = plot_func(*args, **kwargs)
grdevices.dev_off()
if filetype == 'png':
image_display(f'{filename}.{filetype}')
return return_object
def read_pd(file, *args, **kwargs):
if (file.split('.')[-1] == 'txt') or (file.split('.')[-1] == 'tab'):
return pd.read_table(file, header=0, index_col=0, *args, **kwargs)
elif (file.split('.')[-1] == 'xls') or (file.split('.')[-1] == 'xlsx'):
return pd.read_excel(file, *args, **kwargs)
else:
raise IOError("Cannot parse count matrix. Make sure it is .txt, .xls, or .xlsx")
def rout_write(x):
'''
function for setting r_out to print to file instead of jupyter
rpy2.rinterface.set_writeconsole_regular(rout_write)
rpy2.rinterface.set_writeconsole_warnerror(rout_write)
'''
print(x, file=open(f'{os.getcwd()}/R_out_{datetime.now():%Y-%m-%d}.txt', 'a'))
def alert_me(text):
'''
Send me a pop up alert to macosx.
'''
os.system(f'''osascript -e 'tell Application "System Events" to display dialog "{text}"' ''')
def tq_type():
environ = os.environ
if '_' in environ.keys():
jupyter = True if os.environ['_'].endswith('jupyter') else False
elif 'MPLBACKEND' in environ.keys():
jupyter = True if 'ipykernel' in os.environ['MPLBACKEND'] else jupyter
return tqdm_notebook if jupyter else tqdm
def peak_overlap_MC(df_dict, background, permutations=1000, seed=42, notebook=True):
'''
Monte Carlo simulation of peak overlaps in a given background
pvalue calucated as liklihood over emperical random background overlap of shuffled peaks per chromosome.
Inputs
------
df_dict: dictinoary of dataframes in bed format
background genome space: pybedtool bed of background genome space
permutations: number of permutations
seed: random seed
Returns
-------
pvalue
'''
np.random.seed(seed)
tq = tq_type()
# generate probability of chosing a chromosome region based on its size
bregions = background.to_dataframe()
bregions.index = range(len(bregions))
bregions['Size'] = bregions.iloc[:, 2] - bregions.iloc[:, 1]
total_size = bregions.Size.sum()
bregions['fraction'] = bregions.Size / total_size
bed_dict = {name: df.copy() for name, df in df_dict.items()}
# determine length of each peak region
for df in bed_dict.values():
df['Length'] = df.iloc[:, 2] - df.iloc[:, 1]
# determine baseline overlap intersect count of preshuffled peaks.
A, B = bed_dict.values()
overlap = len(BedTool.from_dataframe(A).sort().merge() + BedTool.from_dataframe(B).sort().merge())
results = []
for permutation in tq(range(permutations)):
for df in bed_dict.values():
# randomly pick a region in the background based on size distribution of the regions
index_list = bregions.index.tolist()
df_size = len(df)
bregions_fraction = bregions.fraction
first_pick = np.random.choice(index_list, size=df_size, p=bregions_fraction)
lengths = df.Length.tolist()
alternatives = np.random.choice(index_list, size=df_size, p=bregions_fraction)
# repick regions if the peak length is larger than the region size (this part can be optimized)
regions = []
new_pick = 0
for reg, length in zip(first_pick, lengths):
reg_length = bregions.iloc[reg, 2] - bregions.iloc[reg, 1]
if reg_length > length:
regions.append(reg)
else:
while reg_length <= length:
new_reg = alternatives[new_pick]
reg_length = bregions.iloc[new_reg, 2] - bregions.iloc[new_reg, 1]
new_pick += 1
regions.append(new_reg)
# assign the chromosome
df.iloc[:, 0] = [bregions.iloc[x, 0] for x in regions]
# randomly pick a start within the selected background region within the peak size constraints
df.iloc[:, 1] = [np.random.randint(bregions.iloc[reg, 1], bregions.iloc[reg, 2] - length) for length, reg in zip(lengths, regions)]
# assign end based on peak length
df.iloc[:, 2] = df.iloc[:, 1] + df.Length
new_overlap = len(BedTool.from_dataframe(A).sort().merge() + BedTool.from_dataframe(B).sort().merge())
results.append(1 if new_overlap >= overlap else 0)
p = (sum(results) + 1) / (len(results) + 1)
A_name, B_name = df_dict.keys()
print(f'Number of intersected peaks of {A_name} and {B_name}: {overlap}')
print(f'Number of times simulated intersections exceeded or equaled the actual overlap: {sum(results)}')
print(f'Monte Carlo p-value estimate: {p}')
return p
'''
Implementation of an Enrichr API with graphs
Author: <NAME>
'''
def post_genes(gene_list, description):
'''
posts gene list to Enricr
Returns
-------
dictionary: userListId, shortId
'''
import json
import requests
ENRICHR_URL = 'http://amp.pharm.mssm.edu/Enrichr/addList'
genes_str = '\n'.join([str(x) for x in gene_list])
payload = {'list': (None, genes_str),
'description': (None, description)
}
response = requests.post(ENRICHR_URL, files=payload)
if not response.ok:
raise Exception('Error analyzing gene list')
return json.loads(response.text)
def enrich(userListId, filename, gene_set_library):
'''
Returns
-------
Text file of enrichment results
'''
import requests
ENRICHR_URL = 'http://amp.pharm.mssm.edu/Enrichr/export'
query_string = '?userListId=%s&filename=%s&backgroundType=%s'
url = ENRICHR_URL + query_string % (userListId, filename, gene_set_library)
response = requests.get(url, stream=True)
with open(filename, 'wb') as f:
for chunk in response.iter_content(chunk_size=1024):
if chunk:
f.write(chunk)
return response
def enrichr_barplot(filename, gene_library, out_dir, description, max_n=20,
q_thresh=0.05, color='slategray', display_image=True):
'''
Saves barplot from Enrichr results
Paramaters
----------
filename: enrichr response file
gene_library: gene set library to test
out_dir: result output folder
description: sample or gene set source name
max_n: max number of significant to display
q_thresh: qvalue threshold
color: plot color
dispaly_image: bool
Return
------
None
'''
import seaborn as sns
import matplotlib.pyplot as plt
e_df = pd.read_csv(filename, header=0, sep="\t").sort_values(by=['Adjusted P-value']).head(max_n)
e_df['Clean_term'] = e_df.Term.apply(lambda x: x.split("_")[0])
e_df['log_q'] = -np.log10(e_df['Adjusted P-value'])
plt.clf()
sns.set(context='paper', font='Arial', font_scale=1.2, style='white',
rc={'figure.dpi': 300, 'figure.figsize': (8, 6)}
)
fig, ax = plt.subplots()
fig.suptitle(f'{description} {gene_library.replace("_", " ")} enrichment\n(q<{q_thresh}, max {max_n})')
sig = e_df[e_df['Adjusted P-value'] <= q_thresh].copy()
if len(sig) > 0:
g = sns.barplot(data=sig, x='log_q', y='Clean_term', color=color, ax=ax)
plt.xlabel('q-value (-log$_{10}$)')
plt.ylabel('Enrichment Term')
ymin, ymax = g.get_ylim()
g.vlines(x=-np.log10(q_thresh), ymin=ymin, ymax=ymax, colors='k',
linestyles='dashed', label=f'q = {q_thresh}')
g.legend()
sns.despine()
else:
ax.text(0.5, 0.5, 'No Significant Enrichments.',
horizontalalignment='center',
verticalalignment='center',
transform=ax.transAxes
)
try:
plt.tight_layout(h_pad=1, w_pad=1)
except ValueError:
pass
plt.subplots_adjust(top=0.88)
file = f'{out_dir}{description}_{gene_library}_enrichr.barplot.png'
fig.savefig(file, dpi=300)
plt.close()
image_display(file)
def enrichr(dict_of_genelists, out_dir, dict_of_genelibraries=None, display=True,
q_thresh=0.05, plot_color='slategray', max_n=20 ):
'''
Runs enrichment analysis through Enrichr and plots results
Paramaters
----------
dict_of_genelists: dictionary of description to genelists
dict_of_genelibraries: dictionary of enrichr gene libraries to test against
If None, will use default libraries
display: bool whether to display inline
q_thresh: qvalue threshold
plot_color:
max_n:
'''
out_dir = out_dir if out_dir.endswith('/') else f'{out_dir}/'
gene_libraries ={'KEGG': 'KEGG_2016',
'GO_Biological_Process': 'GO_Biological_Process_2018',
'ChIP-X_Consensus_TFs': 'ENCODE_and_ChEA_Consensus_TFs_from_ChIP-X',
'ChEA': 'ChEA_2016',
'OMIM_Disease': 'OMIM_Disease'
}
libraries = gene_libraries if dict_of_genelibraries is None else dict_of_genelibraries
generator = ((d,g,l,gl) for d,g in dict_of_genelists.items()
for l, gl in libraries.items()
)
for description, genes, library, gene_library in generator:
filename=f'{out_dir}{description}_{library}.enrichr.txt'
post = post_genes(genes, description)
get = enrich(post['userListId'], filename, gene_library)
if get.ok:
enrichr_barplot(filename=filename, gene_library=library, out_dir=out_dir, description=description,
max_n=max_n,q_thresh=q_thresh, color=plot_color, display_image=display)
else:
print(f'Enrichr error: {library}, {description}')
'''
end enrichr
'''
def gsea_dotplot(df_dict, title='', qthresh=0.05, top_term=None, gene_sets=[], dotsize_factor=4, figsize=(4, 10), out_dir='.'):
'''
Makes a dotplot of GSEA results with the dot size as the percent of genes in the leading edge and the color the NES.
Plots only significant dots at given fdr theshold
Inputs
------
df_dict: dictionary of named GSEA results for the analysis. pandas df of gsea_report.xls (use pd.concat to combine pos and neg enrichments)
name: name used for title and filename
qthresh: qvalue theshold for includsion
pgene_sets: list of gene sets to plot. If empty, will plot all with FDR q value < 0.05
top_term: integer specifing top number of sets to plot (by qvalue). None plots all.
dot_size_factor: scale to increase dot size for leading edge %
out_dir: output directory
Returns
-------
Gene_Sets used for plotting
'''
import matplotlib.pyplot as plt
import seaborn as sns
out_dir = val_folder(out_dir)
index = []
# get leading edge percentages
for df in df_dict.values():
if 'NAME' in df.columns.tolist():
df.index = df.NAME
df['le_tags'] = df['LEADING EDGE'].apply(lambda x: x.split('%')[0].split('=')[-1])
df.sort_values(by='NES', ascending=False, inplace=True)
index += df[df['FDR q-val'] < 0.05].index.tolist()
index = list(set(index))
# use gene_sets if provided
if len(gene_sets) > 0:
index = gene_sets
# make master df
data_df = pd.DataFrame()
for name, df in df_dict.items():
df['sample_name'] = name
data_df = | pd.concat([data_df, df.loc[index]]) | pandas.concat |
""" plotting functions for Dataset objects
To Do:
Edit hyp_stats plots to take transitions.HypStats object instead of ioeeg.Dataset object
Remove redundant plotting fns added into EKG classs
Add subsetEEG function to break up concatenated NREM segments for plotting. Will require adjustments
to specified detections added to plot.
"""
import itertools
import igraph as ig
import math
import matplotlib
import matplotlib.dates as mdates
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import shapely.geometry as SG
from matplotlib.widgets import Slider
from pandas.plotting import register_matplotlib_converters
from scipy.signal import find_peaks, butter, sosfiltfilt
from scipy import interpolate
register_matplotlib_converters()
def plotEEG(d, raw=True, filtered=False, spindles=False, spindle_rejects=False):
""" plot multichannel EEG w/ option for double panel raw & filtered. For short, pub-ready
figures. Use vizeeg for data inspection
red = spindle rejects by time domain criteria; dark red = spindle rejects by frequency domain criteria
Parameters
----------
d: instance of ioeeg Dataset class
raw: bool, optional, default: True
Option to plot raw EEG
filtered: bool, optional, default: False
Option to plot filtered EEG
spindles: bool, optional, default: False
Option to plot spindle detections
spindle_rejects: bool, optional, default: False
Option to plot rejected spindle detections
Returns
-------
matplotlib.pyplot figure instance
"""
data = []
title = []
# import data
if raw == True:
raw = d.data
data.append(raw)
title.append('Raw')
if filtered == True:
filtd = d.spindle_calcs.loc(axis=1)[:, 'Filtered']
data.append(filtd)
title.append('Filtered')
# flatten events list by channel for plotting
if spindles == True:
sp_eventsflat = [list(itertools.chain.from_iterable(d.spindle_events[i])) for i in d.spindle_events.keys()]
if spindle_rejects == True:
sp_rej_t_eventsflat = [list(itertools.chain.from_iterable(d.spindle_rejects_t[i])) for i in d.spindle_rejects_t.keys()]
sp_rej_f_eventsflat = [list(itertools.chain.from_iterable(d.spindle_rejects_f[i])) for i in d.spindle_rejects_f.keys()]
# set channels for plotting
channels = [x[0] for x in d.data.columns]
# plot data
fig, axs = plt.subplots(len(data), 1, sharex=True, figsize=(10,10), squeeze=False)
fig.subplots_adjust(hspace=.1, top=.9, bottom=.1, left=.05, right=.95)
for dat, ax, t in zip(data, axs.flatten(), title):
for i, c in enumerate(channels):
# normalize each channel to [0, 1]
dat_ser = pd.Series(dat[(c, t)], index=dat.index)
norm_dat = (dat_ser - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i # subtract i for plotting offset
ax.plot(norm_dat, linewidth=.5, color='C0')
# plot spindles
if spindles == True:
sp_events_TS = [pd.Timestamp(x) for x in sp_eventsflat[i]]
spins = pd.Series(index=norm_dat.index)
spins[sp_events_TS] = norm_dat[sp_events_TS]
ax.plot(spins, color='orange', alpha=0.5)
if spindle_rejects == True:
# plot time-domain rejects
sp_rejs_t_TS = [pd.Timestamp(x) for x in sp_rej_t_eventsflat[i]]
spin_rejects_t = pd.Series(index=norm_dat.index)
spin_rejects_t[sp_rejs_t_TS] = norm_dat[sp_rejs_t_TS]
ax.plot(spin_rejects_t, color='red', alpha=0.5)
# plot frequency-domain rejects
sp_rejs_f_TS = [pd.Timestamp(x) for x in sp_rej_f_eventsflat[i]]
spin_rejects_f = pd.Series(index=norm_dat.index)
spin_rejects_f[sp_rejs_f_TS] = norm_dat[sp_rejs_f_TS]
ax.plot(spin_rejects_f, color='darkred', alpha=0.5)
ax.set_title(t)
ax.set_yticks(list(np.arange(0.5, -(len(channels)-1), -1)))
ax.set_yticklabels(channels)
ax.margins(x=0) # remove white space margins between data and y axis
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
# set overall parameters
fig.suptitle(d.metadata['file_info']['in_num'])
plt.xlabel('Time')
return fig, axs
def plotEEG_singlechan(d, chan, raw=True, filtered=False, rms=False, thresholds=False, spindles=False, spindle_rejects=False):
""" plot single channel EEG. Options for multipaneled calculations. Not for concatenated datasets
Parameters
----------
d: instance of ioeeg Dataset class
chan: str
channel to plot
raw: bool, optional, default: True
Option to plot raw EEG panel
filtered: bool, optional, default: False
Option to plot filtered EEG panel
rms: bool, optional, default: False
Option to plot filtered EEG panel with RMS and RMS moving average
thresholds: bool, optional, default: False
Option to plot spindle threshold lines on rms panel
spindles: bool, optional, default: False
Option to plot filtered EEG with spindle detection panel
spindle_rejects: bool, optional, default: False
Option to plot filtered EEG with spindle rejection panel.
Note: Spindles and spindle_rejects plot on same panel if
both True
Returns
-------
matplotlib.pyplot figure instance
"""
data = []
dtype = []
labels = []
c = chan
# import data
if raw == True:
raw_data = d.data[c, 'Raw']
if filtered == True or rms == True or spindles == True or spindle_rejects == True:
filtd_data = d.spindle_calcs.loc(axis=1)[c, 'Filtered']
# set data to plot
if raw == True:
#raw = d.data[c, 'Raw']
data.append(raw_data)
dtype.append('raw')
labels.append('Raw Signal')
if filtered == True:
#filtd = d.spindle_calcs.loc(axis=1)[c, 'Filtered']
data.append(filtd_data)
dtype.append('filtd')
labels.append('Filtered Signal')
if rms == True:
data.append(filtd_data)
dtype.append('filtd+rms')
labels.append('Filtered Signal')
if spindles == True or spindle_rejects == True:
data.append(filtd_data)
labels.append('Filtered Signal')
if spindles == True and spindle_rejects == False:
dtype.append('filtd+spin')
elif spindles == False and spindle_rejects == True:
dtype.append('filtd+rej')
elif spindles == True and spindle_rejects == True:
dtype.append('filtd+spin+rej')
# pull out thresholds for labels
loSD = d.metadata['spindle_analysis']['sp_loSD']
hiSD = d.metadata['spindle_analysis']['sp_hiSD']
# plot data
fig, axs = plt.subplots(len(data), 1, sharex=True, figsize=(18,6), squeeze=False)
fig.subplots_adjust(hspace=.1, top=.9, bottom=.1, left=.05, right=.95)
for dat, ax, dt, label in zip(data, axs.flatten(), dtype, labels):
# plot EEG
ax.plot(dat, linewidth=.5, color='C0', label=label)
# plot filtered EEG w/ rms & thresholds
if dt == 'filtd+rms':
ax.plot(d.spRMS[c], label='RMS', color='green')
ax.plot(d.spRMSmavg[c], label='RMS moving average', color='orange')
if dt == 'filtd+rms' and thresholds == True:
ax.axhline(d.spThresholds[c].loc['Low Threshold'], linestyle='solid', color='grey', label = f'Mean RMS + {loSD} SD')
ax.axhline(d.spThresholds[c].loc['High Threshold'], linestyle='dashed', color='grey', label = f'Mean RMS + {hiSD} SD')
# plot spindles
if dt =='filtd+spin' or dt =='filtd+spin+rej':
sp_valuesflat = []
sp_eventsflat = []
for n in range(len(d.spindle_events[c])):
for m in range(len(d.spindle_events[c][n])):
sp_valuesflat.append(dat[d.spindle_events[c][n][m]])
sp_eventsflat.append(d.spindle_events[c][n][m])
sp_events_TS = [pd.Timestamp(x) for x in sp_eventsflat]
spins = pd.Series(index=dat.index)
spins[sp_events_TS] = dat[sp_events_TS]
ax.plot(spins, color='orange', alpha=0.5, label='Spindle Detection')
# plot spindle rejections
if dt == 'filtd+rej' or dt == 'filtd+spin+rej':
# plot time-domain rejects
sp_rej_t_valuesflat = []
sp_rej_t_eventsflat = []
for n in range(len(d.spindle_rejects_t[c])):
for m in range(len(d.spindle_rejects_t[c][n])):
sp_rej_t_valuesflat.append(dat[d.spindle_rejects_t[c][n][m]])
sp_rej_t_eventsflat.append(d.spindle_rejects_t[c][n][m])
sp_rej_t_events_TS = [pd.Timestamp(x) for x in sp_rej_t_eventsflat]
spin_rejects_t = pd.Series(index=dat.index)
spin_rejects_t[sp_rej_t_events_TS] = dat[sp_rej_t_events_TS]
ax.plot(spin_rejects_t, color='red', alpha=0.5, label='Rejected Detection (T)')
# plot frequency-domain rejects
sp_rej_f_valuesflat = []
sp_rej_f_eventsflat = []
for n in range(len(d.spindle_rejects_f[c])):
for m in range(len(d.spindle_rejects_f[c][n])):
sp_rej_f_valuesflat.append(dat[d.spindle_rejects_f[c][n][m]])
sp_rej_f_eventsflat.append(d.spindle_rejects_f[c][n][m])
sp_rej_f_events_TS = [pd.Timestamp(x) for x in sp_rej_f_eventsflat]
spin_rejects_f = pd.Series(index=dat.index)
spin_rejects_f[sp_rej_f_events_TS] = dat[sp_rej_f_events_TS]
ax.plot(spin_rejects_f, color='darkred', alpha=0.5, label='Rejected Detection (F)')
ax.legend(loc='lower left')
#ax.set_title(t)
#ax.set_yticks(list(np.arange(0.5, -(len(chan)-1), -1)))
#ax.set_yticklabels(chan)
ax.margins(x=0) # remove white space margins between data and y axis
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
# plot minor axes
seconds = mdates.SecondLocator()
ax.xaxis.set_minor_locator(seconds)
ax.grid(axis='x', which='minor', linestyle=':')
ax.grid(axis='x', which='major')
# set overall parameters
fig.suptitle(d.metadata['file_info']['in_num'])
plt.xlabel('Time')
return fig
def vizeeg(d, raw=True, filtered=False, spindles=False, spindle_rejects=False, slider=True, win_width=15, raw_lowpass=True,
lowpass_freq=25, lowpass_order=4):
""" vizualize multichannel EEG w/ option for double panel raw and/or filtered. Optimized for
inspecting spindle detections (title/axis labels removed for space)
Spindles rejected based on time-domain criteria are plotted in red; rejections based on
frequency-domain criteria are plotted in darkred.
Parameters
----------
d: instance of ioeeg Dataset class
raw: bool, optional, default: True
Option to plot raw EEG
filtered: bool, optional, default: False
Option to plot spindle filtered EEG
spindles: bool, optional, default: False
Option to plot spindle detections
spindle_rejects: bool, optional, default: False
Option to plot rejected spindle detections
slider: bool (default: False)
Option to implement an X-axis slider instead of built-in matplotlib zoom. Useful
for inspecting long segments of EEG with a set window
win_width: int (default: 15)
If using slider option, number of seconds to set window width
raw_lowpass: bool (default: True)
Whether to plot the lowpass filtered raw data [in place of the unchanged raw data]
lowpass_freq: int (default: 25)
Frequency to lowpass the raw data for visualization (if not already applied)
lowpass_order: int (default: 4)
Butterworth lowpass filter order to be used if lowpass_raw is not None (doubles for filtfilt)
Returns
-------
matplotlib.pyplot figure instance
"""
# Set figure size (double height if plotting both raw & filtered)
if raw == True & filtered == True:
figsize = (14, 14)
else:
figsize = (14, 7)
data = []
title = []
# import data
if raw == True:
if not raw_lowpass:
# use the unchanged raw data
raw_data = d.data
elif raw_lowpass:
# use the lowpass filtered raw data
try:
# check if filtered data exists
raw_lowpass_data = d.data_lowpass
except AttributeError:
# apply lowpass filter
d.lowpass_raw(lowpass_freq, lowpass_order)
raw_lowpass_data = d.data_lowpass
if filtered == True:
filtd = d.spindle_calcs.loc(axis=1)[:, 'Filtered']
# set data to plot (title corresponds to multiindex level 2 in data df)
if raw == True:
if not raw_lowpass:
# plot the unchanged data
data.append(raw_data)
title.append('Raw')
elif raw_lowpass:
# plot the lowpass data
data.append(raw_lowpass_data)
title.append('raw_lowpass')
if filtered == True:
data.append(filtd)
title.append('Filtered')
# flatten events list by channel for plotting
if spindles == True:
sp_eventsflat = [list(itertools.chain.from_iterable(d.spindle_events[i])) for i in d.spindle_events.keys()]
if spindle_rejects == True:
# time-domain rejects
sp_rej_t_eventsflat = [list(itertools.chain.from_iterable(d.spindle_rejects_t[i])) for i in d.spindle_rejects_t.keys()]
# frequency domain rejects
sp_rej_f_eventsflat = [list(itertools.chain.from_iterable(d.spindle_rejects_f[i])) for i in d.spindle_rejects_f.keys()]
# set channels for plotting
channels = [x[0] for x in d.data.columns if x[0] not in ['EKG', 'EOG_L', 'EOG_R']]
# set offset multiplier (distance between channels in plot)
mx = 0.1
# plot data
fig, axs = plt.subplots(len(data), 1, sharex=True, figsize=figsize, squeeze=False)
fig.subplots_adjust(hspace=.1, top=.9, bottom=.1, left=.05, right=.95)
yticks = []
for dat, ax, t in zip(data, axs.flatten(), title):
for i, c in enumerate(channels):
# normalize each channel to [0, 1] -> can also simply subtract the mean (cleaner looking), but
# normalization preserves relative differences between channels while putting them on a common scale
dat_ser = pd.Series(dat[(c, t)], index=dat.index)
norm_dat = (dat_ser - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i*mx # subtract i for plotting offset
yticks.append(np.nanmedian(norm_dat))
ax.plot(norm_dat, linewidth=.5, color='C0')
# plot spindles
if spindles == True:
sp_events_TS = [pd.Timestamp(x) for x in sp_eventsflat[i]]
spins = pd.Series(index=norm_dat.index)
spins[sp_events_TS] = norm_dat[sp_events_TS]
ax.plot(spins, color='orange', alpha=0.5)
if spindle_rejects == True:
# plot time-domain rejects
sp_rejs_t_TS = [pd.Timestamp(x) for x in sp_rej_t_eventsflat[i]]
spin_t_rejects = pd.Series(index=norm_dat.index)
spin_t_rejects[sp_rejs_t_TS] = norm_dat[sp_rejs_t_TS]
ax.plot(spin_t_rejects, color='red', alpha=0.5)
# plot frequency-domain rejects
sp_rejs_f_TS = [pd.Timestamp(x) for x in sp_rej_f_eventsflat[i]]
spin_f_rejects = pd.Series(index=norm_dat.index)
spin_f_rejects[sp_rejs_f_TS] = norm_dat[sp_rejs_f_TS]
ax.plot(spin_f_rejects, color='darkred', alpha=0.5)
# remove title to maximize on-screen plot area
#ax.set_title(t)
# set y axis params
ax.set_yticks(yticks)
ax.set_yticklabels(channels)
ax.set_ylim(bottom = yticks[-1]-3*mx, top=yticks[0]+3*mx)
ax.margins(x=0) # remove white space margins between data and y axis
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
# if data roughly 5 mins or less, set minor x-axes
if (d.data.index[-1] - d.data.index[0]).total_seconds() < 400:
seconds = mdates.SecondLocator()
ax.xaxis.set_minor_locator(seconds)
ax.grid(axis='x', which='minor', linestyle=':')
ax.grid(axis='x', which='major')
# set overall parameters
#fig.tight_layout(pad=0) # remove figure padding --> this pushes slider onto fig
# remove labels to maximize on-screen plot area
#plt.xlabel('Time')
#fig.suptitle(d.metadata['file_info']['in_num'])
# option to use x-axis slider insted of matplotlib zoom
if slider:
# plot minor axes --> requires slider for segments longer than 5mins
seconds = mdates.SecondLocator()
ax.xaxis.set_minor_locator(seconds)
ax.grid(axis='x', which='minor', linestyle=':')
ax.grid(axis='x', which='major')
# set initial window
x_min_index = 0
x_max_index = win_width*int(d.s_freq)
x_min = d.data.index[x_min_index]
x_max = d.data.index[x_max_index]
x_dt = x_max - x_min
y_min, y_max = plt.axis()[2], plt.axis()[3]
plt.axis([x_min, x_max, y_min, y_max])
axcolor = 'lightgoldenrodyellow'
axpos = plt.axes([0.2, 0.1, 0.65, 0.03], facecolor=axcolor)
slider_max = len(d.data) - x_max_index - 1
# set slider position object
spos = Slider(axpos, 'Pos', matplotlib.dates.date2num(x_min), matplotlib.dates.date2num(d.data.index[slider_max]))
# format date names
#plt.gcf().autofmt_xdate()
# create slider update function
def update(val):
pos = spos.val
xmin_time = matplotlib.dates.num2date(pos)
xmax_time = matplotlib.dates.num2date(pos) + x_dt
ax.axis([xmin_time, xmax_time, y_min, y_max])
fig.canvas.draw_idle()
# update slider position on click
spos.on_changed(update)
#return fig, axs
return fig
def plotLFP(d, raw=True, filtered=True, thresholds=True, spindles=True, spindle_rejects=True, raw_lowpass=True, lowpass_freq=25,
lowpass_order=4, win_frac=None, xlim=None):
""" plot dual-channel LFP w/ option for double panel raw & filtered.
red = spindle rejects by time domain criteria; dark red = spindle rejects by frequency domain criteria
Parameters
----------
d: instance of ioeeg Dataset class
raw: bool, optional, default: True
Option to plot raw EEG
filtered: bool, optional, default: True
Option to plot filtered EEG
thresholds: bool, optional, default: True
Option to plot spindle detection thresholds
spindles: bool, optional, default: True
Option to plot spindle detections
spindle_rejects: bool, optional, default: True
Option to plot rejected spindle detections
raw_lowpass: bool (default: True)
Whether to plot the lowpass filtered raw data [in place of the unchanged raw data]
lowpass_freq: int (default: 25)
Frequency to lowpass the raw data for visualization (if not already applied)
lowpass_order: int (default: 4)
Butterworth lowpass filter order to be used if lowpass_raw is not None (doubles for filtfilt)
win_frac: str or None (default: None)
window count, if plotting x-axis in windows (ex. '3/4' for window 3 of 4)
xlim: tuple of DateTimeIndex
x-axis values to be used for x-limits
Returns
-------
matplotlib.pyplot figure instance
"""
data = []
title = []
# import data
if raw == True:
if not raw_lowpass:
# use the unchanged raw data
raw_data = d.data
elif raw_lowpass:
# use the lowpass filtered raw data
try:
# check if filtered data exists
raw_lowpass_data = d.data_lowpass
except AttributeError:
# apply lowpass filter
d.lowpass_raw(lowpass_freq, lowpass_order)
raw_lowpass_data = d.data_lowpass
if filtered == True or thresholds == True:
filtd = d.spindle_calcs.loc(axis=1)[:, 'Filtered']
# set data to plot (title corresponds to multiindex level 2 in data df)
if raw == True:
if not raw_lowpass:
# plot the unchanged data
data.append(raw_data)
title.append('Raw')
elif raw_lowpass:
# plot the lowpass data
data.append(raw_lowpass_data)
title.append('raw_lowpass')
if filtered == True:
data.append(filtd)
title.append('Filtered')
if thresholds == True:
data.append(filtd)
title.append('Filtered')
# flatten events list by channel for plotting
if spindles == True:
sp_eventsflat = [list(itertools.chain.from_iterable(d.spindle_events[i])) for i in d.spindle_events.keys()]
if spindle_rejects == True:
sp_rej_t_eventsflat = [list(itertools.chain.from_iterable(d.spindle_rejects_t[i])) for i in d.spindle_rejects_t.keys()]
sp_rej_f_eventsflat = [list(itertools.chain.from_iterable(d.spindle_rejects_f[i])) for i in d.spindle_rejects_f.keys()]
# set channels for plotting
channels = [x[0] for x in d.data.columns]
# plot data
fig, axs = plt.subplots(len(data), 1, sharex=True, figsize=(18,6), squeeze=False)
fig.subplots_adjust(hspace=.2, top=.9, bottom=.1, left=.05, right=.95)
for (e, dat), ax, t in zip(enumerate(data), axs.flatten(), title):
for i, c in enumerate(channels):
# set labels for only the first filtered channel (prevent duplicate legends)
if i == 0:
loSD = d.metadata['spindle_analysis']['sp_loSD']
hiSD = d.metadata['spindle_analysis']['sp_hiSD']
labels = {'RMS': 'RMS', 'RMS mavg': 'RMS mavg', 'lo_thres':f'RMS + {loSD} SD','hi_thres':f'RMS + {hiSD} SD', 'spindles':'Spindle Detection',
'spindle_rejects_t': 'Rejected Detection (time-domain)', 'spindle_rejects_f':'Rejected Detection (frequency-domain)'}
else:
label_keys = ['RMS', 'RMS mavg', 'lo_thres', 'hi_thres', 'spindles', 'spindle_rejects_t', 'spindle_rejects_f']
labels = {k:'_nolegend_' for k in label_keys}
# normalize each channel to [0, 1]; plot signal on 1st & 2nd panels
dat_ser = pd.Series(dat[(c, t)], index=dat.index)
norm_dat = (dat_ser - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i # subtract i for plotting offset
ax.plot(norm_dat, linewidth=.5, color='C0')
# plot thresholds on the second panel
if (thresholds == True) & (e == 1):
# RMS
rms_ser = d.spRMS[c].RMS
norm_rms = (rms_ser - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i
ax.plot(norm_rms, linewidth=.8, color='green', label = labels['RMS'])
# RMS moving average
rmsmavg_ser = d.spRMSmavg[c].RMSmavg
norm_rmsmavg = (rmsmavg_ser - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i
ax.plot(norm_rmsmavg, linewidth=.8, color='orange', label = labels['RMS mavg'])
# threshold values
norm_lo = (d.spThresholds[c].loc['Low Threshold'] - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i
norm_hi = (d.spThresholds[c].loc['High Threshold'] - min(dat_ser))/(max(dat_ser)-min(dat_ser)) - i
ax.axhline(norm_lo, linestyle='solid', color='grey', label = labels['lo_thres'])
ax.axhline(norm_hi, linestyle='dashed', color='grey', label = labels['hi_thres'])
# plot spindles on the 3rd panel
if (spindles == True) & (e == 2):
sp_events_TS = [pd.Timestamp(x) for x in sp_eventsflat[i]]
spins = pd.Series(index=norm_dat.index)
spins[sp_events_TS] = norm_dat[sp_events_TS]
ax.plot(spins, color='orange', alpha=0.5, label=labels['spindles'])
if (spindle_rejects == True) & (e == 2):
# plot time-domain rejects
sp_rejs_t_TS = [ | pd.Timestamp(x) | pandas.Timestamp |
import pandas as pd
import numpy as np
from datetime import datetime
from tqdm import tqdm
from tqdm.notebook import tqdm as tqdmn
try:
from trade import Trade
except:
pass
try:
from backtest.trade import Trade
except:
pass
import chart_studio.plotly as py
import plotly.graph_objs as go
from plotly import subplots
import plotly.express as px
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
pd.options.display.float_format = '{:.5f}'.format
import random
class Backtest:
def __init__(self, strategy, data, from_date, to_date, balance=10000, leverage=0, max_units=10000000, verbose=True, ipynb=False, direct=True, test=False, ddw=0,
commission=0.0, rfr=0.02):
# initial variables
self.strategy = strategy # trading strategy
self.Leverage = leverage # leverage
self.FromDate = str(from_date).split(' ')[0] # starting date
self.ToDate = str(to_date).split(' ')[0] # ending date
self.Data = self.section(data, self.FromDate, self.ToDate) # slice from the dataset
self.Data['MC'] = ((self.Data['AC'] + self.Data['BC']) / 2) # middle close price
self.Data['MO'] = ((self.Data['AO'] + self.Data['BO']) / 2) # middle open price
self.Datasets = [] # all datasets nad instruments
self.Verbose = verbose # verbose checkker
self.ipynb = ipynb # only for Jupyter notebook
self.Direct = direct # calculating instrument units directly or indirectly
self.Test = test # run as a test only, with no balance calculation
self.DDW = ddw # drawdown value
self.RfR = rfr # risk-free rate
# variables for the simulation
self.Commission = commission # commision per trade (percentage)
self.OpenPositions = [] # list of the opened trades
self.CurrentProfit = 0 # unrealized profit/loos
self.GrossLoss = 0 # total loss
self.GrossProfit = 0 # total profit
self.TotalPL = 0 # total profit/loss
self.InitBalance = balance # initial balance
self.Balance = balance # account balance with closed trades
self.MarginLeft = balance # margin left with unrealized profit
self.Unrealized = 0 # unrealized profit/loss
self.MaxUnits = max_units # maximal trading ammount
self.History = [] # list to store previus prices for the user
self.IndicatorList = [] # list to store indicators
columns=['Type', 'Open Time', 'Close Time', 'Units', 'Margin Used', 'Open Price', 'Close Price', 'Spread', 'Profit', 'Balance', 'AutoClose', 'TP', 'SL']
self.Results = pd.DataFrame(columns = ['Ratio', 'Value']) # dataframe for result analysis
self.TradeLog = pd.DataFrame(columns = columns) # pandas dataframe to log activity
self.AutoCloseCount = 0 # counts how many times were trades closed automatically
snp_benchmark = None # loading S&P as benchmark
dji_benchmark = None # loading DJI as benchmark
dax_benchmark = None # loading DAX as benchmark
try:
snp_benchmark = pd.read_csv('data/datasets/spx500usd/spx500usd_hour.csv')
except:
snp_benchmark = pd.read_csv('../data/datasets/spx500usd/spx500usd_hour.csv')
try:
dji_benchmark = pd.read_csv('data/datasets/djiusd/djiusd_hour.csv')
except:
dji_benchmark = pd.read_csv('../data/datasets/djiusd/djiusd_hour.csv')
try:
dax_benchmark = | pd.read_csv('data/datasets/de30eur/de30eur_hour.csv') | pandas.read_csv |
# pylint: disable=E1101
from datetime import datetime
import datetime as dt
import os
import warnings
import nose
import struct
import sys
from distutils.version import LooseVersion
import numpy as np
import pandas as pd
from pandas.compat import iterkeys
from pandas.core.frame import DataFrame, Series
from pandas.core.common import is_categorical_dtype
from pandas.io.parsers import read_csv
from pandas.io.stata import (read_stata, StataReader, InvalidColumnName,
PossiblePrecisionLoss, StataMissingValue)
import pandas.util.testing as tm
from pandas.tslib import NaT
from pandas import compat
class TestStata(tm.TestCase):
def setUp(self):
self.dirpath = tm.get_data_path()
self.dta1_114 = os.path.join(self.dirpath, 'stata1_114.dta')
self.dta1_117 = os.path.join(self.dirpath, 'stata1_117.dta')
self.dta2_113 = os.path.join(self.dirpath, 'stata2_113.dta')
self.dta2_114 = os.path.join(self.dirpath, 'stata2_114.dta')
self.dta2_115 = os.path.join(self.dirpath, 'stata2_115.dta')
self.dta2_117 = os.path.join(self.dirpath, 'stata2_117.dta')
self.dta3_113 = os.path.join(self.dirpath, 'stata3_113.dta')
self.dta3_114 = os.path.join(self.dirpath, 'stata3_114.dta')
self.dta3_115 = os.path.join(self.dirpath, 'stata3_115.dta')
self.dta3_117 = os.path.join(self.dirpath, 'stata3_117.dta')
self.csv3 = os.path.join(self.dirpath, 'stata3.csv')
self.dta4_113 = os.path.join(self.dirpath, 'stata4_113.dta')
self.dta4_114 = os.path.join(self.dirpath, 'stata4_114.dta')
self.dta4_115 = os.path.join(self.dirpath, 'stata4_115.dta')
self.dta4_117 = os.path.join(self.dirpath, 'stata4_117.dta')
self.dta_encoding = os.path.join(self.dirpath, 'stata1_encoding.dta')
self.csv14 = os.path.join(self.dirpath, 'stata5.csv')
self.dta14_113 = os.path.join(self.dirpath, 'stata5_113.dta')
self.dta14_114 = os.path.join(self.dirpath, 'stata5_114.dta')
self.dta14_115 = os.path.join(self.dirpath, 'stata5_115.dta')
self.dta14_117 = os.path.join(self.dirpath, 'stata5_117.dta')
self.csv15 = os.path.join(self.dirpath, 'stata6.csv')
self.dta15_113 = os.path.join(self.dirpath, 'stata6_113.dta')
self.dta15_114 = os.path.join(self.dirpath, 'stata6_114.dta')
self.dta15_115 = os.path.join(self.dirpath, 'stata6_115.dta')
self.dta15_117 = os.path.join(self.dirpath, 'stata6_117.dta')
self.dta16_115 = os.path.join(self.dirpath, 'stata7_115.dta')
self.dta16_117 = os.path.join(self.dirpath, 'stata7_117.dta')
self.dta17_113 = os.path.join(self.dirpath, 'stata8_113.dta')
self.dta17_115 = os.path.join(self.dirpath, 'stata8_115.dta')
self.dta17_117 = os.path.join(self.dirpath, 'stata8_117.dta')
self.dta18_115 = os.path.join(self.dirpath, 'stata9_115.dta')
self.dta18_117 = os.path.join(self.dirpath, 'stata9_117.dta')
self.dta19_115 = os.path.join(self.dirpath, 'stata10_115.dta')
self.dta19_117 = os.path.join(self.dirpath, 'stata10_117.dta')
self.dta20_115 = os.path.join(self.dirpath, 'stata11_115.dta')
self.dta20_117 = os.path.join(self.dirpath, 'stata11_117.dta')
self.dta21_117 = os.path.join(self.dirpath, 'stata12_117.dta')
def read_dta(self, file):
# Legacy default reader configuration
return read_stata(file, convert_dates=True)
def read_csv(self, file):
return read_csv(file, parse_dates=True)
def test_read_empty_dta(self):
empty_ds = | DataFrame(columns=['unit']) | pandas.core.frame.DataFrame |
import pandas as pd
from pandas import DataFrame
import sys
#--------
# Imports medi dataset with icd9 and rxcui descriptions to .csv file
# PARAMETERS:
# medi = medi spreadsheet
# icd9_desc = contains icd9 codes and their descriptions
# rxcui_desc = contains rxcui codes and their descriptions
def add_info_to_medi(medi, icd9_desc, rxcui_desc):
# adding in icd9 descriptions
df_icd9_desc = pd.read_table(icd9_desc, sep=' ', header=None, usecols=[0, 1])
df_icd9_desc.columns = ['ICD9', 'ICD9_DESC']
# adding in rxcui descriptions into the medi spreadsheet
df_rxcui_desc = pd.read_csv(rxcui_desc, encoding='latin-1').drop_duplicates().groupby('RXCUI_IN')['STR'].apply('; '.join)
rxcui_desc = pd.DataFrame({'RXCUI_IN': df_rxcui_desc.index, 'STR': df_rxcui_desc.values})
df_medi = pd.read_csv(medi)
df_medi_desc = pd.merge(df_medi, rxcui_desc, how='left', on='RXCUI_IN')
df_rxcui_icd9 = pd.merge(df_medi_desc, df_icd9_desc, how='left', on='ICD9')
df_rxcui_icd9 = df_rxcui_icd9[['RXCUI_IN', 'STR', 'DRUG_DESC', 'ICD9', 'ICD9_DESC', 'INDICATION_DESCRIPTION', 'MENTIONEDBYRESOURCES',
'HIGHPRECISIONSUBSET', 'POSSIBLE_LABEL_USE']]
df_rxcui_icd9.to_csv('medi_with_icd9_rxcui.csv', index=False)
#--------
# Imports medi_rxcui_icd9 dataset with icd9-phecode mappings to .csv file
# Maps drug (rxcui codes) with clinical phenotype (phecode) through icd9 codes
# PARAMETERS:
# medi_rxcui_icd9 = medi spreadsheet (created from add_info_to_medi function above) with rxcui + icd9 descriptions
# phecode_icd9_mapping = maps phecodes to icd9 codes
def drug_phenotype(phecode_icd9_mapping, medi_rxcui_icd9):
df_rxcui_icd9 = pd.read_csv(medi_rxcui_icd9)
df_phecode_icd9 = pd.read_csv(phecode_icd9_mapping, usecols=['ICD9', 'PheCode'])
result = pd.merge(df_rxcui_icd9, df_phecode_icd9, how='left', on='ICD9').drop_duplicates().sort_values('RXCUI_IN')
result.to_csv('drug_phenotype.csv', index=False)
#print (result)
#--------
# Imports medi_rxcui_icd9 dataset with drug-targeted gene mappings to .csv file
# Maps drugs (rxcui codes) with corresponding targeted genes (HuGOIDs) through unii codes and DrugBank drug IDs
# PARAMETERS:
# unii_rxcui = contains mapping of unii codes to rxcui codes
# unii_drug = contains mapping of unii codes to HuGOIDs (DrugBank), needs to be .txt file
# medi_rxcui_icd9 = medi spreadsheet (created from add_info_to_medi function above) with rxcui + icd9 descriptions
# drug_gene = for each gene, contains list of drugs that target said gene
def drug_gene(unii_rxcui, unii_drug, drug_gene, medi_rxcui_icd9):
df_unii_rxcui = pd.read_csv(unii_rxcui)
df_unii_drug = pd.read_table(unii_drug, header=0, sep=':', usecols=['unii', 'drug_id'])
df_rxcui_icd9 = | pd.read_csv(medi_rxcui_icd9) | pandas.read_csv |
import numpy as np
import pandas as pd
from joblib import Parallel, delayed
from argparse import ArgumentParser
from os import path
from time import time
from utils import trj2blocks
# MDAnalysis
import MDAnalysis as mda
from MDAnalysis.analysis.hydrogenbonds import hbond_analysis
def parse():
'''Parse command line arguments.
Returns:
Namespace object containing input arguments.
'''
parser = ArgumentParser(description='MDTools: Hydrogen bond analysis')
parser.add_argument('-i', '--input', required=True, type=str,
help='Input .xyz file')
parser.add_argument('-n', '--n_cpu', required=True, type=int,
help='Number of CPUs for parallel processing')
parser.add_argument('-c', '--cell_vectors', required=True, type=float,
help='Lattice vectors in angstroms (a, b, c)', nargs=3)
return parser.parse_args()
def hbonds(u, block):
'''Computes hydrogen bond (HB) statistics.
Args:
u: MDAnalysis Universe object containing trajectory.
block: Range of frames composing block.
Returns:
Accepted and donated hydrogen bond counts and surface separations
'''
# Initialize hydrogen bond analysis
hbonds = hbond_analysis.HydrogenBondAnalysis(
u, d_h_a_angle_cutoff=135, d_a_cutoff=3.5)
hbonds.donors_sel = 'name O'
hbonds.acceptors_sel = 'name O'
hbonds.hydrogens_sel = 'name H'
# Run hydrogen bond analysis
hbonds.run(start=block.start, stop=block.stop, verbose=True)
out = hbonds.results.hbonds
# Select oxygen atoms, initialize output arrays
oxygen = u.select_atoms('name O')
acc_counts = np.zeros((len(block), oxygen.n_atoms))
don_counts = np.zeros((len(block), oxygen.n_atoms))
heights = np.zeros((len(block), oxygen.n_atoms))
for i, ts in enumerate(u.trajectory[block.start:block.stop]):
print('Processing blocks %.1f%%' % (100*i/len(block)), end='\r')
# Get all HBs of current frame
step = out[(out[:, 0] == ts.frame)]
# Loop over each oxygen
for j, idx in enumerate(oxygen.indices):
# Get number of accepted and donated HBs + position along z
don_counts[i, j] = len(step[(step[:, 1] == idx)])
acc_counts[i, j] = len(step[(step[:, 3] == idx)])
heights[i, j] = oxygen[j].position[2]
return np.stack((heights, acc_counts, don_counts))
def main():
args = parse()
input = args.input
n_jobs = args.n_cpu
a, b, c = args.cell_vectors
CURRENT_PATH = path.dirname(path.realpath(__file__))
DATA_PATH = path.normpath(path.join(CURRENT_PATH, path.dirname(input)))
base = path.splitext(path.basename(input))[0]
# Initialize universe (time step 0.5 fs)
u = mda.Universe(input, dt=5e-4)
u.add_TopologyAttr('charges')
u.dimensions = np.array([a, b, c, 90, 90, 90])
# Split trajectory into blocks
blocks = trj2blocks.get_blocks(u, n_jobs)
print('Analyzing...')
results = Parallel(n_jobs=n_jobs)(delayed(hbonds)(
u, block) for block in blocks)
# Concatenate results
results = np.concatenate(results, axis=1)
# Save results (heights, accepted HBs, donated HBs) as .csv
df1 = | pd.DataFrame(results[0]) | pandas.DataFrame |
# -*- coding: utf-8 -*-
# pylint: disable
"""
tests.test_validator
~~~~~~~~~~~~~~~~~~~~
:copyright: (c) 2018 by <NAME>.
:license: MIT, see LICENSE for more details.
"""
import numpy as np
from pandas import Series, date_range, to_datetime
from dfmapper import (
DateRangeValidator,
DtypeValidator,
MinValueValidator,
MaxValueValidator,
MaxLengthValidator,
NullableValidator
)
def test_date_range_validator():
series_1 = Series(['2018-01-15 12:00:00', '2018-01-15 13:00:00'])
series_1 = to_datetime(series_1)
date_range_validator_1 = DateRangeValidator(date_range(start='2018-01-01', end='2018-01-30'))
assert date_range_validator_1(series_1) == True
series_2 = Series(['2018-01-15 12:00:00', '2018-01-15 13:00:00'])
series_2 = | to_datetime(series_2) | pandas.to_datetime |
import pandas as pd
from geopy.geocoders import Nominatim
import os
import pathlib as plib
def add_coords(coords, city, state):
gloc = Nominatim(user_agent='my-application', timeout=3)
loc = gloc.geocode(city + ' ' + state)
if loc is None:
coords[city] = None
else:
coords[city] = [loc.raw['lon'], loc.raw['lat']]
pass
def get_coords(cities, state):
coords = {}
for city in cities:
add_coords(coords, city, state)
cts = list(coords.keys())
vals = list(coords.values())
lats = []
lons = []
for x in vals:
if x is None:
lats.append(None)
lons.append(None)
else:
lats.append(x[0])
lons.append(x[1])
df = | pd.DataFrame({'City': cts, 'Latitude': lats, 'Longitude': lons}) | pandas.DataFrame |
'''
PipelineTranscriptDiffExpression.py - Utility functions for
pipeline_transcriptdiffexpression.py
==============================================================
:Author: <NAME>
:Release: $Id$
:Date: |today|
:Tags: Python
Code
----
'''
import cgatpipelines.tasks.expression as Expression
import cgatpipelines.tasks.counts as Counts
import cgatcore.iotools as iotools
from cgatcore import pipeline as P
from cgatcore.pipeline import cluster_runnable
from rpy2.robjects import r as R
import pandas as pd
import numpy as np
import sqlite3
import os
def connect(database, annotations_database):
'''utility function to connect to database.
Use this method to connect to the pipeline database.
Additional databases can be attached here as well.
Returns an sqlite3 database handle.
'''
dbh = sqlite3.connect(database)
statement = '''ATTACH DATABASE '%s' as annotations''' % (
annotations_database)
cc = dbh.cursor()
cc.execute(statement)
cc.close()
return dbh
@cluster_runnable
def runSleuth(design, base_dir, model, contrasts, outfile, counts, tpm,
fdr, lrt=False, reduced_model=None):
''' run sleuth. Note: all samples in the design table must also
have a directory with the same name in `base_dir` with kallisto
results in a file called abundance.h5'''
outfile_prefix = P.snip(outfile, ".tsv")
Design = Expression.ExperimentalDesign(design)
exp = Expression.DEExperiment_Sleuth()
res = exp.run(Design, base_dir, model, contrasts, outfile_prefix,
counts, tpm, fdr, lrt, reduced_model)
res.getResults(fdr)
for contrast in set(res.table['contrast']):
res.plotMA(contrast, outfile_prefix)
res.plotVolcano(contrast, outfile_prefix)
res.table.to_csv(outfile, sep="\t", index=False)
@cluster_runnable
def runSleuthAll(samples, base_dir, counts, tpm):
''' run sleuth for all samples to obtain counts and tpm tables
Note: all samples in the design table must also
have a directory with the same name in `base_dir` with kallisto
results in a file called abundance.h5
'''
design = pd.DataFrame({
"group": ([0, 1] * ((len(samples) + 1) / 2))[0:len(samples)],
"include": [1, ] * len(samples),
"pair": [0, ] * len(samples)})
design.index = samples
Design = Expression.ExperimentalDesign(design)
exp = Expression.DEExperiment_Sleuth()
res = exp.run(Design, base_dir, counts=counts, tpm=tpm,
model="~group", dummy_run=True)
@cluster_runnable
def makeExpressionSummaryPlots(counts_inf, design_inf, logfile):
''' use the plotting methods for Counts object to make summary plots'''
with iotools.openFile(logfile, "w") as log:
plot_prefix = P.snip(logfile, ".log")
# need to manually read in data as index column is not the first column
counts = Counts.Counts(pd.read_table(counts_inf, sep="\t"))
counts.table.set_index(["transcript_id"])
design = Expression.ExperimentalDesign(design_inf)
# make certain counts table only include samples in design
counts.restrict(design)
cor_outfile = plot_prefix + "_pairwise_correlations.png"
pca_var_outfile = plot_prefix + "_pca_variance.png"
pca1_outfile = plot_prefix + "_pc1_pc2.png"
pca2_outfile = plot_prefix + "_pc3_pc4.png"
heatmap_outfile = plot_prefix + "_heatmap.png"
counts_log10 = counts.log(base=10, pseudocount=0.1, inplace=False)
counts_highExp = counts_log10.clone()
counts_highExp.table['order'] = counts_highExp.table.apply(
np.mean, axis=1)
counts_highExp.table.sort(["order"], ascending=0, inplace=True)
counts_highExp.table = counts_highExp.table.iloc[0:500, :]
counts_highExp.table.drop("order", axis=1, inplace=True)
log.write("plot correlations: %s\n" % cor_outfile)
counts_log10.plotPairwiseCorrelations(cor_outfile, subset=1000)
log.write("plot pc3,pc4: %s\n" % pca1_outfile)
counts_log10.plotPCA(design,
pca_var_outfile, pca1_outfile,
x_axis="PC1", y_axis="PC2",
colour="group", shape="group")
log.write("plot pc3,pc4: %s\n" % pca2_outfile)
counts_log10.plotPCA(design,
pca_var_outfile, pca2_outfile,
x_axis="PC3", y_axis="PC4",
colour="group", shape="group")
log.write("plot heatmap: %s\n" % heatmap_outfile)
counts_highExp.heatmap(heatmap_outfile)
@cluster_runnable
def identifyLowConfidenceTranscripts(infile, outfile):
''' identify transcripts which cannot be confidently quantified in
the simulation '''
df = pd.read_table(infile, sep="\t", index_col=0)
with iotools.openFile(outfile, "w") as outf:
outf.write("%s\t%s\n" % ("transcript_id", "reason"))
# identify transcript with low fraction of kmers - these show
# poorer correlation between ground truth and esimated counts
low_fraction = df[df['fraction_bin'] < 0.03].index.tolist()
for transcript in low_fraction:
outf.write("%s\t%s\n" % (transcript, "low_kmers"))
# identify transcript with poor accuracy of quantification
low_accuracy = df[[abs(x) > 0.585 for x in
df['log2diff_tpm']]].index.tolist()
for transcript in low_accuracy:
outf.write("%s\t%s\n" % (transcript, "poor_accuracy"))
@cluster_runnable
def mergeAbundanceCounts(infile, outfile, counts):
''' merge the abundance and simulation counts files for
each simulation '''
df_abund = pd.read_table(infile, sep="\t", index_col=0)
df_counts = pd.read_table(counts, sep="\t", index_col=0)
df_abund.columns = [x if x != "tpm" else "est_tpm"
for x in df_abund.columns]
df_merge = pd.merge(df_abund, df_counts, left_index=True, right_index=True)
df_merge.index.name = "id"
df_merge.to_csv(outfile, sep="\t")
@cluster_runnable
def calculateCorrelations(infiles, outfile, bin_step=1):
''' calculate correlation across simulation iterations per transcript'''
abund, kmers = infiles
df_abund = pd.read_table(abund, sep="\t", index_col=0)
df_kmer = pd.read_table(kmers, sep="\t", index_col=0)
# this is hacky, it's doing all against all correlations for the
# two columns and subsetting
df_agg_tpm = df_abund.groupby(level=0)[[
"est_tpm", "tpm"]].corr().ix[0::2, 'tpm']
# drop the "read_count" level, make into dataframe and rename column
df_agg_tpm.index = df_agg_tpm.index.droplevel(1)
df_agg_tpm = pd.DataFrame(df_agg_tpm)
df_agg_tpm.columns = ["tpm_cor"]
df_agg_count = df_abund.groupby(level=0)[[
"est_counts", "read_count"]].corr().ix[0::2, 'read_count']
# drop the "read_count" level, make into dataframe and rename column
df_agg_count.index = df_agg_count.index.droplevel(1)
df_agg_count = pd.DataFrame(df_agg_count)
df_agg_count.columns = ["counts_cor"]
# merge and bin the unique fraction values
df_agg = pd.merge(df_agg_count, df_agg_tpm,
left_index=True, right_index=True)
df_final = | pd.merge(df_kmer, df_agg, left_index=True, right_index=True) | pandas.merge |
"""Transform signaling data to smoothed trajectories."""
import sys
import numpy
import pandas as pd
import geopandas as gpd
import shapely.geometry
import matplotlib.patches
import matplotlib.pyplot as plt
import mobilib.voronoi
SAMPLING = pd.Timedelta('00:01:00')
STD = pd.Timedelta('00:05:00')
def smoothen(array, std_quant):
return pd.Series(array).rolling(
int(numpy.ceil(8 * std_quant)),
min_periods=0,
center=True,
win_type='gaussian'
).mean(std=std_quant)
def trajectory(df, xcol, ycol, sampling, std):
ts = pd.date_range(df.index.min(), df.index.max(), freq=sampling)
obs_ind = ts.searchsorted(df.index)
xs_src = numpy.full(ts.size, numpy.nan)
xs_src[obs_ind] = df[xcol]
ys_src = numpy.full(ts.size, numpy.nan)
ys_src[obs_ind] = df[ycol]
std_quant = std / sampling
return smoothen(xs_src, std_quant), smoothen(ys_src, std_quant), ts
if __name__ == '__main__':
signals = pd.read_csv(sys.argv[1], sep=';')
signals = signals[signals['phone_nr'] == int(sys.argv[3])]
signals['pos_time'] = | pd.to_datetime(signals['pos_time']) | pandas.to_datetime |
""" Plots the tracker charts. """
import os
from datetime import datetime
from datetime import timedelta
import logging
import shutil
import pathlib
import multiprocessing as mp
import typing
from timeit import default_timer as timer
import pandas as pd
import numpy as np
from scipy.interpolate import interp1d
import plotly.express as px
import plotly.graph_objects as go
SCRIPT_DIR = os.path.dirname(os.path.abspath(__file__))
CHART_OUTPUT = os.path.join(SCRIPT_DIR, "_includes", "tracker", "charts")
TEMPLATE = 'plotly_dark'
PERIOD_DAYS = [14, 30]
MA_SUFFIX = '_MA7'
MA_NAME = "7-day MA"
REGION = 'Region'
CITY_MUN = 'CityMunRes'
CASE_REP_TYPE = 'CaseRepType'
ONSET_PROXY = 'OnsetProxy'
RECOVER_PROXY = 'RecoverProxy'
CASE_STATUS = 'CaseStatus'
DATE_CLOSED = 'DateClosed'
AGE_GROUP_CATEGORYARRAY=['0 to 4', '5 to 9', '10 to 14', '15 to 19', '20 to 24',
'25 to 29', '30 to 34', '35 to 39', '40 to 44', '45 to 49', '50 to 54',
'55 to 59', '60 to 64', '65 to 69', '70 to 74', '75 to 79', '80+', 'No Data'
]
# Number of processes to launch when applying a parallel processing.
# We leave one core idle to avoid hogging all the resources.
num_processes = 1 if (mp.cpu_count() <= 2) else mp.cpu_count() - 1
def apply_parallel(df: pd.DataFrame, func, n_proc=num_processes):
""" Apply function to the dataframe using multiprocessing.
The initial plan was to use modin but because there are still a lot of
missing features and instability in modin, I've resorted to doing the
parallel processing in here.
"""
logging.info(f"Running multiprocessing on {func.__name__} with {n_proc} processes")
df_split = np.array_split(df, n_proc)
pool = mp.Pool(n_proc)
df = pd.concat(pool.map(func, df_split))
pool.close()
pool.join()
return df
def write_table(header, body, filename):
logging.info(f"Writing {filename}")
table = "".join(f"<th>{cell}</th>" for cell in header)
for row in body:
row_html = "".join(f"<td>{cell}</td>" for cell in row)
table += f"<tr>{row_html}</tr>"
table = f"<div><table>{table}</table></div>"
with open(f"{CHART_OUTPUT}/{filename}.html", 'w') as f:
f.write(table)
def write_chart(fig, filename):
logging.info(f"Writing {filename}")
fig.update_layout(template=TEMPLATE)
fig.update_layout(margin=dict(l=5, r=5, b=50, t=70))
fig.write_html(f"{CHART_OUTPUT}/{filename}.html", include_plotlyjs='cdn',
full_html=False)
def plot_for_period(df: pd.DataFrame,
plot: typing.Callable,
filter_df: typing.Callable[[pd.DataFrame, int], pd.DataFrame],
**kwargs):
"""Execute the plot function for the overall data and for each PERIOD_DAYS.
The plot function must take a 'write_chart' keyword argument which is the
function that writes the chart to a file.
"""
plot(df, **kwargs)
for days in PERIOD_DAYS:
filtered = filter_df(df, days)
kwargs_passed = kwargs.copy()
# Append the period in days at the end of the filename.
if 'write_chart' in kwargs_passed:
write_fn = kwargs_passed['write_chart']
else:
write_fn = write_chart
kwargs_passed['write_chart'] = (lambda fig, filename :
write_fn(fig, f"{filename}{days}days"))
plot(df, **kwargs_passed)
def filter_date_range(data, start=None, end=None, date_column=None):
"""Return only the rows within the specified date range."""
if date_column:
if start and end:
return data[(data[date_column] >= start) & (data[date_column] <= end)]
elif start:
return data[data[date_column] >= start]
elif end:
return data[data[date_column] <= end]
else:
raise ValueError("Either start or end should not be None")
else:
if start and end:
return data[(data.index >= start) & (data.index <= end)]
elif start:
return data[data.index >= start]
elif end:
return data[data.index <= end]
else:
raise ValueError("Either start or end should not be None")
def filter_latest(data, days, date_column=None, return_latest=True):
""" Filter data by the days indicated in the days parameter.
If date_column is None, the index is used as the date column.
The default behavior is to return the latest data. If return_latest is
False, the latest data is filtered out instead.
"""
if date_column:
cutoff_date = data[date_column].max() - pd.Timedelta(days=days)
logging.debug(f"Filtering {date_column} cutoff {cutoff_date}.")
if return_latest:
return data[data[date_column] > cutoff_date]
return data[data[date_column] < cutoff_date]
else:
cutoff_date = data.index.max()- | pd.Timedelta(days=days) | pandas.Timedelta |
import unittest
import numpy as np
import pandas as pd
from sklearn.cluster import DBSCAN, KMeans
from sklearn.covariance import EmpiricalCovariance, MinCovDet
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.mixture import GaussianMixture
from dsbox.ml.outliers import CovarianceOutliers, GaussianProcessOutliers
from dsbox.ml.outliers import GMMOutliers, ClusteringOutliers
from dsbox.ml.outliers import KMeansOneClusterOutliers, KMeansIterativeOneClusterOutliers
from dsbox.ml.outliers import MADOutliers, FFTOutliers
class CovarianceOutliersTest(unittest.TestCase):
def test_covarianceoutliers_constructor_should_accept_different_scikit_covariance_estimators(self):
# given
robust_cov = MinCovDet()
emp_cov = EmpiricalCovariance()
# when
cov_outliers_1 = CovarianceOutliers(emp_cov)
cov_outliers_2 = CovarianceOutliers(robust_cov)
# then
self.assertTrue(isinstance(cov_outliers_1, CovarianceOutliers))
self.assertTrue(isinstance(cov_outliers_2, CovarianceOutliers))
def test_covarianceoutliers_predict_proba_gives_biggest_proba_to_biggest_outlier(self):
# given
df = pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1])
# when
cov_outliers = CovarianceOutliers()
cov_outliers.fit(df)
probas = cov_outliers.predict_proba(df)
outlier_index = np.argmax(probas)
# then
outlier_index_true = 6
self.assertEqual(outlier_index_true, outlier_index)
def test_covarianceoutliers_predict_should_return_correct_values(self):
# given
df = pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1])
# when
cov_outliers = CovarianceOutliers()
cov_outliers.fit(df)
outliers = cov_outliers.predict(df)
# then
outliers_true = [False, False, False, False, False, False, True, True, False, False, False,
False, False, False]
self.assertListEqual(outliers_true, outliers.tolist())
class GaussianProcessOutliersTest(unittest.TestCase):
def test_gpoutliers_predict_should_return_correct_values(self):
# given
data = np.random.random_sample(1000) * 2 - 1
data[300] = 5
data[700] = -6
df = pd.DataFrame(data)
# when
gp_outliers = GaussianProcessOutliers(GaussianProcessRegressor(alpha=0.9, normalize_y=True), n_samples=100)
gp_outliers.fit(df)
outliers = gp_outliers.predict(df, confidence=0.999)
# then
outlier_positions_true = [300, 700]
self.assertTrue(outliers[outlier_positions_true[0]])
self.assertTrue(outliers[outlier_positions_true[1]])
class KMeansOneClusterOutliersTest(unittest.TestCase):
def test_kmeansonecluster_outliers_predict_should_return_correct_values(self):
# given
df = pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1])
# when
kmoc_outliers = KMeansOneClusterOutliers()
kmoc_outliers.fit(df)
outliers = kmoc_outliers.predict(df)
# then
outliers_true = [False, False, False, False, False, False, True, True, False, False, False,
False, False, False]
self.assertListEqual(outliers_true, outliers.tolist())
class KMeansIterativeOneClusterOutliersTest(unittest.TestCase):
def test_kmeans_iterative_onecluster_outliers_predict_should_return_correct_values(self):
# given
df = pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1])
# when
kmoc_outliers = KMeansIterativeOneClusterOutliers()
outliers = kmoc_outliers.fit_predict(df)
# then
outliers_true = [2., 1., 1., 2., 2., -1., 0., 0., 1., 1., -1., 1., 1., 2.]
self.assertListEqual(outliers_true, outliers.tolist())
class GMMOutliersTest(unittest.TestCase):
def test_gmm_outliers_predict_should_return_correct_values(self):
# given
df = pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1])
# when
gmm_outliers = GMMOutliers()
gmm_outliers.fit(df)
outliers = gmm_outliers.predict(df)
# then
outliers_true = [False, False, False, False, False, False, True, True, False, False, False,
False, False, False]
self.assertListEqual(outliers_true, outliers.tolist())
class ClusteringOutliersTest(unittest.TestCase):
def test_clustering_outliers_predict_proba_with_unclustered_strategy_should_return_correct_values(self):
# given
df = pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1])
# when
clustering_outliers = ClusteringOutliers(cluster_estimator=DBSCAN(min_samples=2), strategy='unclustered')
clustering_outliers.fit(df)
outliers = clustering_outliers.predict_proba(df)
# then
outliers_true = [0., 0., 0., 0., 1., 0., 1., 1., 1., 1., 0., 0.,
1., 0.]
self.assertListEqual(outliers_true, outliers.tolist())
def test_clustering_outliers_predict_with_unclustered_strategy_should_return_correct_values(self):
# given
df = | pd.DataFrame([1, 0, 0, 1, 10, 2, 115, 110, 32, 16, 2, 0, 15, 1]) | pandas.DataFrame |
# https://www.udemy.com/course/ai-finance
import os
from glob import glob
from datetime import datetime, date
import random
import pandas as pd
import yfinance as yf
def load_stock_list(market='br', symbols_list='', qty = 100):
"""This function loads the desirable symbols.
Args:
market (str): accepts only 'br' or 'us'
symbols_list (list): list of symbols.
Returns:
list: list of desirable symbols.
"""
qty = qty - len(symbols_list)
symbols = | pd.read_csv('./data/interim/lst_stock_symbols.txt', sep=';') | pandas.read_csv |
import os
import numpy as np
import pandas as pd
from pkg_resources import resource_filename
def load_arrests(return_X_y=False, give_pandas=False):
"""
Loads the arrests dataset which can serve as a benchmark for fairness. It is data on
the police treatment of individuals arrested in Toronto for simple possession of small
quantities of marijuana. The goal is to predict whether or not the arrestee was released
with a summons while maintaining a degree of fairness.
:param return_X_y: If True, returns ``(data, target)`` instead of a dict object.
:param give_pandas: give the pandas dataframe instead of X, y matrices (default=False)
:Example:
>>> from sklego.datasets import load_arrests
>>> X, y = load_arrests(return_X_y=True)
>>> X.shape
(5226, 7)
>>> y.shape
(5226,)
>>> load_arrests(give_pandas=True).columns
Index(['released', 'colour', 'year', 'age', 'sex', 'employed', 'citizen',
'checks'],
dtype='object')
The dataset was copied from the carData R package and can originally be found in:
- Personal communication from <NAME>, York University.
The documentation page of the dataset from the package can be viewed here:
http://vincentarelbundock.github.io/Rdatasets/doc/carData/Arrests.html
"""
filepath = resource_filename("sklego", os.path.join("data", "arrests.zip"))
df = pd.read_csv(filepath)
if give_pandas:
return df
X, y = (
df[["colour", "year", "age", "sex", "employed", "citizen", "checks"]].values,
df["released"].values,
)
if return_X_y:
return X, y
return {"data": X, "target": y}
def load_chicken(return_X_y=False, give_pandas=False):
"""
Loads the chicken dataset. The chicken data has 578 rows and 4 columns
from an experiment on the effect of diet on early growth of chicks.
The body weights of the chicks were measured at birth and every second
day thereafter until day 20. They were also measured on day 21.
There were four groups on chicks on different protein diets.
:param return_X_y: If True, returns ``(data, target)`` instead of a dict object.
:param give_pandas: give the pandas dataframe instead of X, y matrices (default=False)
:Example:
>>> from sklego.datasets import load_chicken
>>> X, y = load_chicken(return_X_y=True)
>>> X.shape
(578, 3)
>>> y.shape
(578,)
>>> load_chicken(give_pandas=True).columns
Index(['weight', 'time', 'chick', 'diet'], dtype='object')
The datasets can be found in the following sources:
- Crowder, M. and <NAME>. (1990), Analysis of Repeated Measures, Chapman and Hall (example 5.3)
- Hand, D. and <NAME>. (1996), Practical Longitudinal Data Analysis, Chapman and Hall (table A.2)
"""
filepath = resource_filename("sklego", os.path.join("data", "chickweight.zip"))
df = pd.read_csv(filepath)
if give_pandas:
return df
if give_pandas:
return df
X, y = df[["time", "diet", "chick"]].values, df["weight"].values
if return_X_y:
return X, y
return {"data": X, "target": y}
def load_abalone(return_X_y=False, give_pandas=False):
"""
Loads the abalone dataset where the goal is to predict the gender of the creature.
:param return_X_y: If True, returns ``(data, target)`` instead of a dict object.
:param give_pandas: give the pandas dataframe instead of X, y matrices (default=False)
:Example:
>>> from sklego.datasets import load_abalone
>>> X, y = load_abalone(return_X_y=True)
>>> X.shape
(4177, 8)
>>> y.shape
(4177,)
>>> load_abalone(give_pandas=True).columns
Index(['sex', 'length', 'diameter', 'height', 'whole_weight', 'shucked_weight',
'viscera_weight', 'shell_weight', 'rings'],
dtype='object')
The dataset was copied from Kaggle and can originally be found in: can be found in the following sources:
- <NAME>, <NAME>, <NAME>, <NAME> and <NAME> (1994)
"The Population Biology of Abalone (_Haliotis_ species) in Tasmania."
Sea Fisheries Division, Technical Report No. 48 (ISSN 1034-3288)
"""
filepath = resource_filename("sklego", os.path.join("data", "abalone.zip"))
df = pd.read_csv(filepath)
if give_pandas:
return df
X = df[
[
"length",
"diameter",
"height",
"whole_weight",
"shucked_weight",
"viscera_weight",
"shell_weight",
"rings",
]
].values
y = df["sex"].values
if return_X_y:
return X, y
return {"data": X, "target": y}
def load_heroes(return_X_y=False, give_pandas=False):
"""
A dataset from a video game: "heroes of the storm". The goal of the dataset
is to predict the attack type. Note that the pandas dataset returns more information.
This is because we wanted to keep the X simple in the return_X_y case.
:param return_X_y: If True, returns ``(data, target)`` instead of a dict object.
:param give_pandas: give the pandas dataframe instead of X, y matrices (default=False)
:Example:
>>> X, y = load_heroes(return_X_y=True)
>>> X.shape
(84, 2)
>>> y.shape
(84,)
>>> df = load_heroes(give_pandas=True)
>>> df.columns
Index(['name', 'attack_type', 'role', 'health', 'attack', 'attack_spd'], dtype='object')
"""
filepath = resource_filename("sklego", os.path.join("data", "heroes.zip"))
df = pd.read_csv(filepath)
if give_pandas:
return df
X = df[["health", "attack"]].values
y = df["attack_type"].values
if return_X_y:
return X, y
return {"data": X, "target": y}
def make_simpleseries(
n_samples=365 * 5,
trend=0.001,
season_trend=0.001,
noise=0.5,
give_pandas=False,
seed=None,
stack_noise=False,
start_date=None,
):
"""
Generate a very simple timeseries dataset to play with. The generator
assumes to generate daily data with a season, trend and noise.
:param n_samples: The number of days to simulate the timeseries for.
:param trend: The long term trend in the dataset.
:param season_trend: The long term trend in the seasonality.
:param noise: The noise that is applied to the dataset.
:param give_pandas: Return a pandas dataframe instead of a numpy array.
:param seed: The seed value for the randomness.
:param stack_noise: Set the noise to be stacked by a cumulative sum.
:param start_date: Also add a start date (only works if `give_pandas`=True).
:return: numpy array unless dataframe is specified
:Example:
>>> from sklego.datasets import make_simpleseries
>>> make_simpleseries(seed=42)
array([-0.34078806, -0.61828731, -0.18458236, ..., -0.27547402,
-0.38237413, 0.13489355])
>>> make_simpleseries(give_pandas=True, start_date="2018-01-01", seed=42).head(3)
yt date
0 -0.340788 2018-01-01
1 -0.618287 2018-01-02
2 -0.184582 2018-01-03
"""
if seed:
np.random.seed(seed)
time = np.arange(0, n_samples)
noise = np.random.normal(0, noise, n_samples)
if stack_noise:
noise = noise.cumsum()
r1, r2 = np.random.normal(0, 1, 2)
seasonality = r1 * np.sin(time / 365 * 2 * np.pi) + r2 * np.cos(
time / 365 * 4 * np.pi + 1
)
result = seasonality + season_trend * seasonality * time + trend * time + noise
if give_pandas:
if start_date:
stamps = pd.date_range(start_date, periods=n_samples)
return | pd.DataFrame({"yt": result, "date": stamps}) | pandas.DataFrame |
import argparse
import os
import os.path as path
import pandas as pd
import cv2
import progressbar
from annotations import ImageAnnotation, SUPPORTED_CLASSES
from keypoints_detection.KeypointDetector import KeypointDetector
from keypoints_detection.factory import create_keypoint_detector
allowed_extensions = ['.jpg']
class InvalidDirectoryStructureError(Exception):
pass
def parse_args():
ap = argparse.ArgumentParser(description='Processes images placed in images_directory with keypoint detector and '
'creates annotation file for whole dataset in csv format. Images in '
'images_directory must be organized in subfolders named by image '
'class names')
ap.add_argument("-i", "--images_directory", required=True,
help="Path to directory with images")
ap.add_argument("-m", "--model_path", required=True,
help="Path to directory with images")
return ap.parse_args()
def get_images_to_process(directory_path):
image_path_to_class_mapping = {}
for subdirectory_name in list_directories(directory_path):
subdirectory_path = os.path.join(directory_path, subdirectory_name)
class_name = subdirectory_name
if not os.path.isdir(subdirectory_path):
raise InvalidDirectoryStructureError(
"Incorrect directory structure. images_directory should contain "
"images in subdirectories named by class name")
if class_name in SUPPORTED_CLASSES:
for file_path in list_files_with_extension(subdirectory_path, allowed_extensions):
if image_path_to_class_mapping.get(file_path) is not None:
print(f'Duplicate file {file_path}')
continue
image_path_to_class_mapping[file_path] = class_name
return image_path_to_class_mapping
def process_images(images_directory, keypoint_detector: KeypointDetector):
annotations_df = pd.DataFrame()
images_to_class_mapping = get_images_to_process(images_directory)
progress_bar = progressbar.ProgressBar(max_value=len(images_to_class_mapping))
for i, image_info in enumerate(images_to_class_mapping.items()):
image_path, class_name = image_info
keypoints = process_image(image_path, keypoint_detector)
annotations_df = annotations_df.append(
get_image_annotation(image_path, class_name, keypoints), ignore_index=True)
progress_bar.update(i)
return annotations_df
def process_image(image_path, keypoint_detector: KeypointDetector):
keypoints = keypoint_detector.detect(image_path)
if len(keypoints) == 0:
return None
# We process only first detection
# TODO: Select detection by size or something else
keypoints = keypoints[0]
return keypoints
def get_image_annotation(image_path, class_name, keypoints):
if keypoints is None:
return | pd.DataFrame() | pandas.DataFrame |
# Copyright 2021 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import account
import allocation
import investment
import mock
import pandas as pd
import unittest
import portfolio
class BuildPortolioTest(unittest.TestCase):
def setUp(self):
self._account_desc = """
[
{
"institution": "Fidelity",
"name": "<NAME>",
"filename": "Personal_Account_Positions.csv",
"taxable": "True",
"headers": {
"name": "Account Name/Number",
"symbol": "Symbol",
"description": "Description",
"num_shares": "Quantity",
"share_price": "Last Price"
}
}
]
"""
@mock.patch('account.Account')
def test_build_portfolio(self, mock_account):
test_account = mock.MagicMock(account.Account, autospect=True)
test_account.name = '<NAME>'
test_account.institution = 'Fidelity'
test_account.account_file = 'Personal_Account_Positions.csv'
test_account.is_taxable = True
test_account.holdings = [
investment.Investment('CRISX', 'Small Cap Value Fund Inst', 'GOOGLE LLC 401(K) SAVINGS PLAN', 18576.337,
share_price=18.36)]
test_account.options = [holding.fund for holding in test_account.holdings]
mock_account.return_value = test_account
with mock.patch('portfolio.open', mock.mock_open(read_data=self._account_desc)) as m:
actual_portfolio = portfolio.build_portfolio('data/accounts.json')
expected_portfolio = portfolio.Portfolio([test_account])
self.assertEqual(actual_portfolio, expected_portfolio)
class PortfolioTest(unittest.TestCase):
def setUp(self):
fidelity_account = mock.MagicMock(account.Account, autospect=True)
fidelity_account.name = 'ACME 401(K) SAVINGS PLAN'
fidelity_account.institution = 'Fidelity'
fidelity_account.account_file = 'Fidelity_Positions.csv'
fidelity_account.is_taxable = True
fidelity_account.holdings = [
investment.Investment('CRISX', investment.AssetClass.SMALL_CAP, 'Small Cap Value Fund Class Institutional',
500, share_price=10),
investment.Investment('FSKAX', investment.AssetClass.CORE_US, 'Fidelity Total Market Index', 1000,
share_price=10),
investment.Investment('CASH', investment.AssetClass.CASH, 'Money Market', 5000,
share_price=1)
]
fidelity_account.options = [holding.fund for holding in fidelity_account.holdings]
vanguard_account = mock.MagicMock(account.Account, autospec=True)
vanguard_account.name = '<NAME>'
vanguard_account.institution = 'Vanguard'
vanguard_account.account_file = 'Vanguard_Positions.csv'
vanguard_account.is_taxable = False
vanguard_account.holdings = [
investment.Investment('VNQ', investment.AssetClass.REAL_ESTATE, 'Vanguard Real Estate Index Fund', 2000,
share_price=10)
]
vanguard_account.options = [holding.fund for holding in vanguard_account.holdings]
self._portfolio = portfolio.Portfolio([fidelity_account, vanguard_account])
def test_get_allocation_by_asset_class(self):
allocation_index = pd.Index([investment.AssetClass.CORE_US, investment.AssetClass.SMALL_CAP,
investment.AssetClass.CASH, investment.AssetClass.REAL_ESTATE], name='asset_class')
expected_allocation_df = pd.DataFrame(data=[10000.0, 5000.0, 5000.0, 20000.0], index=allocation_index,
columns=['value'])
expected_allocation_df.sort_index(inplace=True)
pd.testing.assert_frame_equal(self._portfolio.get_allocation_by_asset_class().sort_index(inplace=False),
expected_allocation_df)
def test_get_allocation_by_institution(self):
allocation_index = | pd.Index(['Fidelity', 'Vanguard'], name='institution') | pandas.Index |
"""
Transforms the extracted data.
"""
import click
import feather
import numpy as np
import pandas as pd
import re
import tqdm
import yaml
from logging import *
from typing import *
def yes_no(x: str) -> float:
"""
Transforms a yes/no value to a numeric value.
Args:
x: The value to transform.
Returns:
The transformed value.
Raises:
ValueError: When the value cannot be translated.
"""
if pd.isnull(x) or x == '':
return np.nan
y = x.lower()
if y in ['y', 'pos']:
return 1.
elif y.startswith('n'):
return 0.
raise ValueError('cannot translate yes/no value: {!r}'.format(x))
def float_(x: str) -> float:
"""
Transforms what should be a float value to a numeric value.
Args:
x: The value to transform.
Returns:
The transformed value.
Raises:
ValueError: When the value cannot be translated.
"""
if pd.isnull(x):
return np.nan
try:
return float(x)
except ValueError:
y = x.lower()
if y.startswith('neg'):
return 0.
m = re.match(r'^(\d+)', y)
if m is not None:
return float(m.group(1))
m = re.match(r'^<(.+)$', y)
if m is not None:
return float(m.group(1)) - 1e-6
m = re.match(r'^>(.+)$', y)
if m is not None:
return float(m.group(1)) + 1e-6
raise ValueError('cannot parse float value: {!r}'.format(x))
def sex_female(x: str) -> float:
"""
Transforms a sex into a numeric value denoting whether a patient is
female.
Args:
x: The value to transform.
Returns:
The transformed value.
Raises:
ValueError: When the value cannot be translated.
"""
if | pd.isnull(x) | pandas.isnull |
"""Tests for Table Schema integration."""
import json
from collections import OrderedDict
import numpy as np
import pandas as pd
import pytest
from pandas import DataFrame
from pandas.core.dtypes.dtypes import (
PeriodDtype, CategoricalDtype, DatetimeTZDtype)
from pandas.io.json.table_schema import (
as_json_table_type,
build_table_schema,
make_field,
set_default_names)
class TestBuildSchema(object):
def setup_method(self, method):
self.df = DataFrame(
{'A': [1, 2, 3, 4],
'B': ['a', 'b', 'c', 'c'],
'C': pd.date_range('2016-01-01', freq='d', periods=4),
'D': pd.timedelta_range('1H', periods=4, freq='T'),
},
index=pd.Index(range(4), name='idx'))
def test_build_table_schema(self):
result = build_table_schema(self.df, version=False)
expected = {
'fields': [{'name': 'idx', 'type': 'integer'},
{'name': 'A', 'type': 'integer'},
{'name': 'B', 'type': 'string'},
{'name': 'C', 'type': 'datetime'},
{'name': 'D', 'type': 'duration'},
],
'primaryKey': ['idx']
}
assert result == expected
result = build_table_schema(self.df)
assert "pandas_version" in result
def test_series(self):
s = pd.Series([1, 2, 3], name='foo')
result = build_table_schema(s, version=False)
expected = {'fields': [{'name': 'index', 'type': 'integer'},
{'name': 'foo', 'type': 'integer'}],
'primaryKey': ['index']}
assert result == expected
result = build_table_schema(s)
assert 'pandas_version' in result
def test_series_unnamed(self):
result = build_table_schema(pd.Series([1, 2, 3]), version=False)
expected = {'fields': [{'name': 'index', 'type': 'integer'},
{'name': 'values', 'type': 'integer'}],
'primaryKey': ['index']}
assert result == expected
def test_multiindex(self):
df = self.df.copy()
idx = pd.MultiIndex.from_product([('a', 'b'), (1, 2)])
df.index = idx
result = build_table_schema(df, version=False)
expected = {
'fields': [{'name': 'level_0', 'type': 'string'},
{'name': 'level_1', 'type': 'integer'},
{'name': 'A', 'type': 'integer'},
{'name': 'B', 'type': 'string'},
{'name': 'C', 'type': 'datetime'},
{'name': 'D', 'type': 'duration'},
],
'primaryKey': ['level_0', 'level_1']
}
assert result == expected
df.index.names = ['idx0', None]
expected['fields'][0]['name'] = 'idx0'
expected['primaryKey'] = ['idx0', 'level_1']
result = build_table_schema(df, version=False)
assert result == expected
class TestTableSchemaType(object):
def test_as_json_table_type_int_data(self):
int_data = [1, 2, 3]
int_types = [np.int, np.int16, np.int32, np.int64]
for t in int_types:
assert as_json_table_type(np.array(
int_data, dtype=t)) == 'integer'
def test_as_json_table_type_float_data(self):
float_data = [1., 2., 3.]
float_types = [np.float, np.float16, np.float32, np.float64]
for t in float_types:
assert as_json_table_type(np.array(
float_data, dtype=t)) == 'number'
def test_as_json_table_type_bool_data(self):
bool_data = [True, False]
bool_types = [bool, np.bool]
for t in bool_types:
assert as_json_table_type(np.array(
bool_data, dtype=t)) == 'boolean'
def test_as_json_table_type_date_data(self):
date_data = [pd.to_datetime(['2016']),
pd.to_datetime(['2016'], utc=True),
pd.Series(pd.to_datetime(['2016'])),
pd.Series(pd.to_datetime(['2016'], utc=True)),
pd.period_range('2016', freq='A', periods=3)]
for arr in date_data:
assert as_json_table_type(arr) == 'datetime'
def test_as_json_table_type_string_data(self):
strings = [pd.Series(['a', 'b']), pd.Index(['a', 'b'])]
for t in strings:
assert as_json_table_type(t) == 'string'
def test_as_json_table_type_categorical_data(self):
assert as_json_table_type(pd.Categorical(['a'])) == 'any'
assert as_json_table_type(pd.Categorical([1])) == 'any'
assert as_json_table_type(pd.Series(pd.Categorical([1]))) == 'any'
assert as_json_table_type(pd.CategoricalIndex([1])) == 'any'
assert as_json_table_type(pd.Categorical([1])) == 'any'
# ------
# dtypes
# ------
def test_as_json_table_type_int_dtypes(self):
integers = [np.int, np.int16, np.int32, np.int64]
for t in integers:
assert as_json_table_type(t) == 'integer'
def test_as_json_table_type_float_dtypes(self):
floats = [np.float, np.float16, np.float32, np.float64]
for t in floats:
assert as_json_table_type(t) == 'number'
def test_as_json_table_type_bool_dtypes(self):
bools = [bool, np.bool]
for t in bools:
assert as_json_table_type(t) == 'boolean'
def test_as_json_table_type_date_dtypes(self):
# TODO: datedate.date? datetime.time?
dates = [np.datetime64, np.dtype("<M8[ns]"), PeriodDtype(),
DatetimeTZDtype('ns', 'US/Central')]
for t in dates:
assert as_json_table_type(t) == 'datetime'
def test_as_json_table_type_timedelta_dtypes(self):
durations = [np.timedelta64, np.dtype("<m8[ns]")]
for t in durations:
assert as_json_table_type(t) == 'duration'
def test_as_json_table_type_string_dtypes(self):
strings = [object] # TODO
for t in strings:
assert as_json_table_type(t) == 'string'
def test_as_json_table_type_categorical_dtypes(self):
# TODO: I think before is_categorical_dtype(Categorical)
# returned True, but now it's False. Figure out why or
# if it matters
assert as_json_table_type(pd.Categorical(['a'])) == 'any'
assert as_json_table_type(CategoricalDtype()) == 'any'
class TestTableOrient(object):
def setup_method(self, method):
self.df = DataFrame(
{'A': [1, 2, 3, 4],
'B': ['a', 'b', 'c', 'c'],
'C': pd.date_range('2016-01-01', freq='d', periods=4),
'D': pd.timedelta_range('1H', periods=4, freq='T'),
'E': pd.Series(pd.Categorical(['a', 'b', 'c', 'c'])),
'F': pd.Series(pd.Categorical(['a', 'b', 'c', 'c'],
ordered=True)),
'G': [1., 2., 3, 4.],
'H': pd.date_range('2016-01-01', freq='d', periods=4,
tz='US/Central'),
},
index=pd.Index(range(4), name='idx'))
def test_build_series(self):
s = pd.Series([1, 2], name='a')
s.index.name = 'id'
result = s.to_json(orient='table', date_format='iso')
result = json.loads(result, object_pairs_hook=OrderedDict)
assert "pandas_version" in result['schema']
result['schema'].pop('pandas_version')
fields = [{'name': 'id', 'type': 'integer'},
{'name': 'a', 'type': 'integer'}]
schema = {
'fields': fields,
'primaryKey': ['id'],
}
expected = OrderedDict([
('schema', schema),
('data', [OrderedDict([('id', 0), ('a', 1)]),
OrderedDict([('id', 1), ('a', 2)])])])
assert result == expected
def test_to_json(self):
df = self.df.copy()
df.index.name = 'idx'
result = df.to_json(orient='table', date_format='iso')
result = json.loads(result, object_pairs_hook=OrderedDict)
assert "pandas_version" in result['schema']
result['schema'].pop('pandas_version')
fields = [
{'name': 'idx', 'type': 'integer'},
{'name': 'A', 'type': 'integer'},
{'name': 'B', 'type': 'string'},
{'name': 'C', 'type': 'datetime'},
{'name': 'D', 'type': 'duration'},
{'constraints': {'enum': ['a', 'b', 'c']},
'name': 'E',
'ordered': False,
'type': 'any'},
{'constraints': {'enum': ['a', 'b', 'c']},
'name': 'F',
'ordered': True,
'type': 'any'},
{'name': 'G', 'type': 'number'},
{'name': 'H', 'type': 'datetime', 'tz': 'US/Central'}
]
schema = {
'fields': fields,
'primaryKey': ['idx'],
}
data = [
OrderedDict([('idx', 0), ('A', 1), ('B', 'a'),
('C', '2016-01-01T00:00:00.000Z'),
('D', 'P0DT1H0M0S'),
('E', 'a'), ('F', 'a'), ('G', 1.),
('H', '2016-01-01T06:00:00.000Z')
]),
OrderedDict([('idx', 1), ('A', 2), ('B', 'b'),
('C', '2016-01-02T00:00:00.000Z'),
('D', 'P0DT1H1M0S'),
('E', 'b'), ('F', 'b'), ('G', 2.),
('H', '2016-01-02T06:00:00.000Z')
]),
OrderedDict([('idx', 2), ('A', 3), ('B', 'c'),
('C', '2016-01-03T00:00:00.000Z'),
('D', 'P0DT1H2M0S'),
('E', 'c'), ('F', 'c'), ('G', 3.),
('H', '2016-01-03T06:00:00.000Z')
]),
OrderedDict([('idx', 3), ('A', 4), ('B', 'c'),
('C', '2016-01-04T00:00:00.000Z'),
('D', 'P0DT1H3M0S'),
('E', 'c'), ('F', 'c'), ('G', 4.),
('H', '2016-01-04T06:00:00.000Z')
]),
]
expected = OrderedDict([('schema', schema), ('data', data)])
assert result == expected
def test_to_json_float_index(self):
data = pd.Series(1, index=[1., 2.])
result = data.to_json(orient='table', date_format='iso')
result = json.loads(result, object_pairs_hook=OrderedDict)
result['schema'].pop('pandas_version')
expected = (
OrderedDict([('schema', {
'fields': [{'name': 'index', 'type': 'number'},
{'name': 'values', 'type': 'integer'}],
'primaryKey': ['index']
}),
('data', [OrderedDict([('index', 1.0), ('values', 1)]),
OrderedDict([('index', 2.0), ('values', 1)])])])
)
assert result == expected
def test_to_json_period_index(self):
idx = pd.period_range('2016', freq='Q-JAN', periods=2)
data = pd.Series(1, idx)
result = data.to_json(orient='table', date_format='iso')
result = json.loads(result, object_pairs_hook=OrderedDict)
result['schema'].pop('pandas_version')
fields = [{'freq': 'Q-JAN', 'name': 'index', 'type': 'datetime'},
{'name': 'values', 'type': 'integer'}]
schema = {'fields': fields, 'primaryKey': ['index']}
data = [OrderedDict([('index', '2015-11-01T00:00:00.000Z'),
('values', 1)]),
OrderedDict([('index', '2016-02-01T00:00:00.000Z'),
('values', 1)])]
expected = OrderedDict([('schema', schema), ('data', data)])
assert result == expected
def test_to_json_categorical_index(self):
data = pd.Series(1, pd.CategoricalIndex(['a', 'b']))
result = data.to_json(orient='table', date_format='iso')
result = json.loads(result, object_pairs_hook=OrderedDict)
result['schema'].pop('pandas_version')
expected = (
OrderedDict([('schema',
{'fields': [{'name': 'index', 'type': 'any',
'constraints': {'enum': ['a', 'b']},
'ordered': False},
{'name': 'values', 'type': 'integer'}],
'primaryKey': ['index']}),
('data', [
OrderedDict([('index', 'a'),
('values', 1)]),
OrderedDict([('index', 'b'), ('values', 1)])])])
)
assert result == expected
def test_date_format_raises(self):
with pytest.raises(ValueError):
self.df.to_json(orient='table', date_format='epoch')
# others work
self.df.to_json(orient='table', date_format='iso')
self.df.to_json(orient='table')
def test_make_field_int(self):
data = [1, 2, 3]
kinds = [pd.Series(data, name='name'), pd.Index(data, name='name')]
for kind in kinds:
result = make_field(kind)
expected = {"name": "name", "type": 'integer'}
assert result == expected
def test_make_field_float(self):
data = [1., 2., 3.]
kinds = [pd.Series(data, name='name'), pd.Index(data, name='name')]
for kind in kinds:
result = make_field(kind)
expected = {"name": "name", "type": 'number'}
assert result == expected
def test_make_field_datetime(self):
data = [1., 2., 3.]
kinds = [pd.Series(pd.to_datetime(data), name='values'),
pd.to_datetime(data)]
for kind in kinds:
result = make_field(kind)
expected = {"name": "values", "type": 'datetime'}
assert result == expected
kinds = [pd.Series(pd.to_datetime(data, utc=True), name='values'),
pd.to_datetime(data, utc=True)]
for kind in kinds:
result = make_field(kind)
expected = {"name": "values", "type": 'datetime', "tz": "UTC"}
assert result == expected
arr = pd.period_range('2016', freq='A-DEC', periods=4)
result = | make_field(arr) | pandas.io.json.table_schema.make_field |
import pandas as pd
p1 = | pd.Series({'a':10,'b':20,'c':30}) | pandas.Series |
import csv
import pandas as pd
import numpy as np
######=================================================########
###### Segment A.1 ########
######=================================================########
SimDays = 365
SimHours = SimDays * 24
HorizonHours = 24 ##planning horizon (e.g., 24, 48, 72 hours etc.)
TransLoss = 0.075 ##transmission loss as a percent of generation
n1criterion = 0.75 ##maximum line-usage as a percent of line-capacity
res_margin = 0.15 ##minimum reserve as a percent of system demand
spin_margin = 0.50 ##minimum spinning reserve as a percent of total reserve
data_name = 'pownet_data_camb_2016'
######=================================================########
###### Segment A.2 ########
######=================================================########
#read parameters for dispatchable resources (coal/gas/oil/biomass generators, imports)
df_gen = | pd.read_csv('data_camb_genparams.csv',header=0) | pandas.read_csv |
#!/usr/bin/env python
# coding: utf-8
# In[1]:
import os
from pathlib import Path
testfolder = str(Path().resolve().parent.parent / 'PV_ICE' / 'TEMP' / 'ElectricFutures')
# Another option using relative address; for some operative systems you might need '/' instead of '\'
# testfolder = os.path.abspath(r'..\..\PV_DEMICE\TEMP')
print ("Your simulation will be stored in %s" % testfolder)
# In[2]:
if not os.path.exists(testfolder):
os.makedirs(testfolder)
# In[3]:
MATERIALS = ['glass','silver','silicon', 'copper','aluminium_frames']
MATERIAL = MATERIALS[0]
MODULEBASELINE = r'..\..\baselines\ElectrificationFutures_2021\EF-CapacityByState-basecase.csv'
MODULEBASELINE_High = r'..\..\baselines\ElectrificationFutures_2021\EF-CapacityByState-LowREHighElec.csv'
# In[4]:
import PV_ICE
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# In[5]:
PV_ICE.__version__
# ### Loading Module Baseline. Will be used later to populate all the columsn otehr than 'new_Installed_Capacity_[MW]' which will be supplied by the REEDS model
# In[6]:
r1 = PV_ICE.Simulation(name='Simulation1', path=testfolder)
#r1.createScenario(name='US', file=r'..\..\baselines\ReedsSubset\baseline_modules_US_Reeds_EF.csv')
r1.createScenario(name='US', file=r'..\..\baselines\ElectrificationFutures_2021\baseline_modules_US_NREL_Electrification_Futures_2021_basecase.csv')
baseline = r1.scenario['US'].data
baseline = baseline.drop(columns=['new_Installed_Capacity_[MW]'])
baseline.set_index('year', inplace=True)
baseline.index = pd.PeriodIndex(baseline.index, freq='A') # A -- Annual
baseline.head()
# In[7]:
plt.rcParams.update({'font.size': 22})
plt.rcParams['figure.figsize'] = (12, 5)
# In[8]:
df = pd.read_csv(MODULEBASELINE)
df.set_index(['Type','State','year'], inplace=True)
df.head()
# In[9]:
for ii in range (len(df.unstack(level=2))):
STATE = df.unstack(level=2).iloc[ii].name[1]
SCEN = df.unstack(level=2).iloc[ii].name[0]
SCEN=SCEN.replace('+', '_')
filetitle = 'base_'+SCEN+'_'+STATE +'.csv'
subtestfolder = os.path.join(testfolder, 'baselines')
if not os.path.exists(subtestfolder):
os.makedirs(subtestfolder)
filetitle = os.path.join(subtestfolder, filetitle)
A = df.unstack(level=2).iloc[ii]
A = A.droplevel(level=0)
A.name = 'new_Installed_Capacity_[MW]'
A = pd.DataFrame(A)
A.index=pd.PeriodIndex(A.index, freq='A')
A = pd.DataFrame(A)
A['new_Installed_Capacity_[MW]'] = A['new_Installed_Capacity_[MW]'] # 0.85 marketshares['Si'] already included
# Add other columns
A = pd.concat([A, baseline.reindex(A.index)], axis=1)
header = "year,new_Installed_Capacity_[MW],mod_eff,mod_reliability_t50,mod_reliability_t90," "mod_degradation,mod_lifetime,mod_MFG_eff,mod_EOL_collection_eff,mod_EOL_collected_recycled," "mod_Repair,mod_MerchantTail,mod_Reuse\n" "year,MW,%,years,years,%,years,%,%,%,%,%,%\n"
with open(filetitle, 'w', newline='') as ict:
# Write the header lines, including the index variable for
# the last one if you're letting Pandas produce that for you.
# (see above).
for line in header:
ict.write(line)
# savedata.to_csv(ict, index=False)
A.to_csv(ict, header=False)
# In[10]:
df = pd.read_csv(MODULEBASELINE_High)
df.set_index(['Type','State','year'], inplace=True)
df.head()
# In[11]:
for ii in range (len(df.unstack(level=2))):
STATE = df.unstack(level=2).iloc[ii].name[1]
SCEN = df.unstack(level=2).iloc[ii].name[0]
SCEN=SCEN.replace('+', '_')
subtestfolder = os.path.join(testfolder, 'baselines')
if not os.path.exists(subtestfolder):
os.makedirs(subtestfolder)
filetitle = 'LowREHighElec_'+SCEN+'_'+STATE +'.csv'
filetitle = os.path.join(subtestfolder, filetitle)
A = df.unstack(level=2).iloc[ii]
A = A.droplevel(level=0)
A.name = 'new_Installed_Capacity_[MW]'
A = pd.DataFrame(A)
A.index=pd.PeriodIndex(A.index, freq='A')
A = pd.DataFrame(A)
A['new_Installed_Capacity_[MW]'] = A['new_Installed_Capacity_[MW]'] # 0.85 marketshares['Si'] already included
# Add other columns
A = pd.concat([A, baseline.reindex(A.index)], axis=1)
header = "year,new_Installed_Capacity_[MW],mod_eff,mod_reliability_t50,mod_reliability_t90," "mod_degradation,mod_lifetime,mod_MFG_eff,mod_EOL_collection_eff,mod_EOL_collected_recycled," "mod_Repair,mod_MerchantTail,mod_Reuse\n" "year,MW,%,years,years,%,years,%,%,%,%,%\n"
with open(filetitle, 'w', newline='') as ict:
# Write the header lines, including the index variable for
# the last one if you're letting Pandas produce that for you.
# (see above).
for line in header:
ict.write(line)
# savedata.to_csv(ict, index=False)
A.to_csv(ict, header=False)
# In[12]:
ict
# # DO SIMULATIONS
# In[13]:
Sims=['base','LowREHighElec']
# In[14]:
baselineinstalls = os.path.join(testfolder, 'baselines')
onlyfiles = [f for f in os.listdir(baselineinstalls)]
# In[15]:
sim1 = [f for f in onlyfiles if f.startswith(Sims[0])]
sim2 = [f for f in onlyfiles if f.startswith(Sims[1])]
# In[16]:
STATEs = [i.split('_', 4)[2] for i in sim1]
STATEs = [i.split('.', 1)[0] for i in STATEs]
Type = [i.split('_', 4)[1] for i in sim1]
# In[31]:
#for ii in range (0, 1): #len(scenarios):
i = 0
r1 = PV_ICE.Simulation(name=Sims[i], path=testfolder)
for jj in range (0, len(sim1)):
filetitle = sim1[jj]
filetitle = os.path.join(testfolder, 'baselines', filetitle)
scen = Type[jj]+'_'+STATEs[jj]
r1.createScenario(name=scen, file=filetitle)
r1.scenario[scen].addMaterial('glass', file=r'..\..\baselines\SolarFutures_2021\baseline_material_glass_Reeds.csv')
r1.scenario[scen].addMaterial('silicon', file=r'..\..\baselines\SolarFutures_2021\baseline_material_silicon_Reeds.csv')
r1.scenario[scen].addMaterial('silver', file=r'..\..\baselines\SolarFutures_2021\baseline_material_silver_Reeds.csv')
r1.scenario[scen].addMaterial('copper', file=r'..\..\baselines\SolarFutures_2021\baseline_material_copper_Reeds.csv')
r1.scenario[scen].addMaterial('aluminum', file=r'..\..\baselines\SolarFutures_2021\baseline_material_aluminium_Reeds.csv')
r1.scenario[scen].material['glass'].materialdata = r1.scenario[scen].material['glass'].materialdata[8:].reset_index(drop=True)
r1.scenario[scen].material['silicon'].materialdata = r1.scenario[scen].material['silicon'].materialdata[8:].reset_index(drop=True)
r1.scenario[scen].material['silver'].materialdata = r1.scenario[scen].material['silver'].materialdata[8:].reset_index(drop=True)
r1.scenario[scen].material['copper'].materialdata = r1.scenario[scen].material['copper'].materialdata[8:].reset_index(drop=True)
r1.scenario[scen].material['aluminum'].materialdata = r1.scenario[scen].material['aluminum'].materialdata[8:].reset_index(drop=True)
i = 1
r2 = PV_ICE.Simulation(name=Sims[i], path=testfolder)
for jj in range (0, len(STATEs)):
filetitle = sim2[jj]
filetitle = os.path.join(testfolder, 'baselines', filetitle)
scen = Type[jj]+'_'+STATEs[jj]
r2.createScenario(name=scen, file=filetitle)
r2.scenario[scen].addMaterial('glass', file=r'..\..\baselines\SolarFutures_2021\baseline_material_glass_Reeds.csv')
r2.scenario[scen].addMaterial('silicon', file=r'..\..\baselines\SolarFutures_2021\baseline_material_silicon_Reeds.csv')
r2.scenario[scen].addMaterial('silver', file=r'..\..\baselines\SolarFutures_2021\baseline_material_silver_Reeds.csv')
r2.scenario[scen].addMaterial('copper', file=r'..\..\baselines\SolarFutures_2021\baseline_material_copper_Reeds.csv')
r2.scenario[scen].addMaterial('aluminum', file=r'..\..\baselines\SolarFutures_2021\baseline_material_aluminium_Reeds.csv')
r2.scenario[scen].material['glass'].materialdata = r2.scenario[scen].material['glass'].materialdata[8:].reset_index(drop=True)
r2.scenario[scen].material['silicon'].materialdata = r2.scenario[scen].material['silicon'].materialdata[8:].reset_index(drop=True)
r2.scenario[scen].material['silver'].materialdata = r2.scenario[scen].material['silver'].materialdata[8:].reset_index(drop=True)
r2.scenario[scen].material['copper'].materialdata = r2.scenario[scen].material['copper'].materialdata[8:].reset_index(drop=True)
r2.scenario[scen].material['aluminum'].materialdata = r2.scenario[scen].material['aluminum'].materialdata[8:].reset_index(drop=True)
# In[33]:
IRENA= False
PERFECTMFG = True
mats = ['glass', 'silicon','silver','copper','aluminum']
ELorRL = 'EL'
if IRENA:
if ELorRL == 'RL':
weibullInputParams = {'alpha': 5.3759, 'beta':30} # Regular-loss scenario IRENA
if ELorRL == 'EL':
weibullInputParams = {'alpha': 2.49, 'beta':30} # Regular-loss scenario IRENA
if PERFECTMFG:
for jj in range (0, len(r1.scenario.keys())):
scen = Type[jj]+'_'+STATEs[jj]
r1.scenario[scen].data['mod_lifetime'] = 40
r1.scenario[scen].data['mod_MFG_eff'] = 100.0
r2.scenario[scen].data['mod_lifetime'] = 40
r2.scenario[scen].data['mod_MFG_eff'] = 100.0
for kk in range(0, len(mats)):
mat = mats[kk]
r1.scenario[scen].material[mat].materialdata['mat_MFG_eff'] = 100.0
r2.scenario[scen].material[mat].materialdata['mat_MFG_eff'] = 100.0
r1.calculateMassFlow(weibullInputParams=weibullInputParams)
r2.calculateMassFlow(weibullInputParams=weibullInputParams)
title_Method = 'Irena_'+ELorRL
else:
r1.calculateMassFlow()
r2.calculateMassFlow()
title_Method = 'PVICE'
# # OPEN EI
# In[34]:
kk=0
SFScenarios = [r1, r2]
SFScenarios[kk].name
# In[35]:
# WORK ON THIS FOIR OPENEI
keyw=['mat_Virgin_Stock','mat_Total_EOL_Landfilled','mat_Total_MFG_Landfilled', 'mat_Total_Landfilled',
'new_Installed_Capacity_[MW]','Installed_Capacity_[W]']
keywprint = ['VirginMaterialDemand','EOLMaterial', 'ManufacturingScrap','ManufacturingScrapAndEOLMaterial',
'NewInstalledCapacity','InstalledCapacity']
keywunits = ['MetricTonnes', 'MetricTonnes', 'MetricTonnes', 'MetricTonnes',
'MW','MW']
keywdcumneed = [True,True,True,True,
True,False]
keywdlevel = ['material','material','material','material',
'module','module']
keywscale = [1000000, 1000000, 1000000, 1000000,
1,1e6]
materials = ['glass', 'silicon', 'silver', 'copper', 'aluminum']
# Loop over SF Scenarios
scenariolist = pd.DataFrame()
for kk in range(0, len(SFScenarios)):
# Loop over Materials
for zz in range (0, len(STATEs)):
foo = | pd.DataFrame() | pandas.DataFrame |
# Copyright (c) 2017, Intel Research and Development Ireland Ltd.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
__authors__ = '<NAME>, <NAME>'
__copyright__ = "Copyright (c) 2017, Intel Research and Development Ireland Ltd."
__license__ = "Apache 2.0"
__maintainer__ = "<NAME>"
__email__ = "<EMAIL>"
__status__ = "Development"
from analytics_engine.heuristics.beans.infograph import InfoGraphNode, InfoGraphNodeType, \
InfoGraphNodeCategory, InfoGraphNodeLayer
import pandas
import math
class Fingerprint(object):
# Deprecated
@staticmethod
def _node_is_nic_on_management_net(node, graph, mng_net_name):
node_name = InfoGraphNode.get_name(node)
node_type = InfoGraphNode.get_type(node)
if node_type == InfoGraphNodeType.VIRTUAL_NIC or \
node_type == InfoGraphNodeType.VIRTUAL_NIC_2:
neighs = graph.neighbors(node_name)
for n in neighs:
neighbor = InfoGraphNode.\
get_node(graph, n)
if InfoGraphNode.get_type(neighbor) == \
InfoGraphNodeType.VIRTUAL_NETWORK:
network_name = \
InfoGraphNode.get_attributes(
neighbor)['name']
if network_name == mng_net_name:
return True
return False
@staticmethod
def workload_capacity_usage(annotated_subgraph):
"""
This is a type of fingerprint
"""
# TODO: Validate graph
categories = list()
categories.append(InfoGraphNodeCategory.COMPUTE)
categories.append(InfoGraphNodeCategory.NETWORK)
# TODO: Add a Volume to the workloads to get HD usage
categories.append(InfoGraphNodeCategory.STORAGE)
# TODO: Get telemetry for Memory
categories.append(InfoGraphNodeCategory.MEMORY)
fingerprint = dict()
counter = dict()
for category in categories:
fingerprint[category] = 0
counter[category] = 0
# calculation of the fingerprint on top of the virtual resources
local_subgraph = annotated_subgraph.copy()
local_subgraph.filter_nodes('layer', "physical")
local_subgraph.filter_nodes('layer', "service")
for node in local_subgraph.nodes(data=True):
# if Fingerprint._node_is_nic_on_management_net(
# node, annotated_subgraph, mng_net_name):
# continue
category = InfoGraphNode.get_category(node)
utilization = InfoGraphNode.get_utilization(node)
if 'utilization' in utilization.columns.values:
mean = utilization['utilization'].mean()
fingerprint[category] += mean
counter[category] += 1
# This is just an average
# TODO: Improve the average
for category in categories:
if counter[category] > 0:
fingerprint[category] = \
fingerprint[category] / counter[category]
return fingerprint
@staticmethod
def machine_capacity_usage(annotated_subgraph):
"""
This is a type of fingerprint from the infrastructure perspective
"""
# TODO: Validate graph
categories = list()
categories.append(InfoGraphNodeCategory.COMPUTE)
categories.append(InfoGraphNodeCategory.NETWORK)
# TODO: Add a Volume to the workloads to get HD usage
categories.append(InfoGraphNodeCategory.STORAGE)
# TODO: Get telemetry for Memory
categories.append(InfoGraphNodeCategory.MEMORY)
fingerprint = dict()
counter = dict()
for category in categories:
fingerprint[category] = 0
counter[category] = 0
# calculation of the fingerprint on top of the virtual resources
local_subgraph = annotated_subgraph.copy()
local_subgraph.filter_nodes('layer', "virtual")
local_subgraph.filter_nodes('layer', "service")
local_subgraph.filter_nodes('type', 'machine')
for node in local_subgraph.nodes(data=True):
# if Fingerprint._node_is_nic_on_management_net(
# node, annotated_subgraph, mng_net_name):
# continue
name = InfoGraphNode.get_name(node)
category = InfoGraphNode.get_category(node)
utilization = InfoGraphNode.get_utilization(node)
if 'utilization' in utilization.columns.values:
# LOG.info("NODE: {} - CATEGORY: {}".format(name, category))
mean = utilization['utilization'].mean()
fingerprint[category] += mean
counter[category] += 1
# This is just an average
# TODO: Improve the average
for category in categories:
if counter[category] > 0:
fingerprint[category] = \
fingerprint[category] / counter[category]
return fingerprint
@staticmethod
def compute_node(annotated_subgraph, hostname=None):
"""
This is a type of fingerprint from the infrastructure perspective
"""
# TODO: Validate graph
data = dict()
statistics = dict()
compute = InfoGraphNodeCategory.COMPUTE
data[compute] = pandas.DataFrame()
statistics[compute] = {'mean': 0, 'median': 0, 'min': 0, 'max': 0, 'var': 0, 'std_dev': 0}
network = InfoGraphNodeCategory.NETWORK
data[network] = pandas.DataFrame()
statistics[network] = {'mean': 0, 'median': 0, 'min': 0, 'max': 0, 'var': 0, 'std_dev': 0}
storage = InfoGraphNodeCategory.STORAGE
data[storage] = | pandas.DataFrame() | pandas.DataFrame |
import wiggum as wg
from itertools import combinations
import pandas as pd
import sys
import logging
from sklearn import mixture
import numpy as np
import json
def updateMetaData(labeled_df, meta):
"""
Update Meta Data
Parameters
-----------
labeled_df : DataFrame
LabeledDataFrame
meta : DataFrame
data organized in a pandas dataframe
Returns
--------
None
"""
meta_list =json.loads(meta)
meta_df_user = pd.DataFrame(meta_list)
# set var_type from user input
var_types = meta_df_user['var_type'].tolist()
labeled_df.set_var_types(var_types)
# set isCount from user input
roles = meta_df_user['role'].tolist()
labeled_df.set_roles(roles)
# set roles from user input
meta_df_user['isCount'] = meta_df_user['isCount'].replace({'Y': True, 'N': False})
counts = meta_df_user['isCount'].tolist()
labeled_df.set_counts(counts)
# set weighting_var from user input
meta_df_user['weighting_var'] = meta_df_user['weighting_var'].replace('N/A', np.nan)
weighting_vars = meta_df_user['weighting_var'].tolist()
labeled_df.set_weighting_vars(weighting_vars)
return labeled_df
def checkSameMetadata(labeled_df, meta):
"""
Check if any metadata changes
Parameters
-----------
labeled_df : DataFrame
LabeledDataFrame
meta : DataFrame
user input metadata
Returns
--------
checkResult : Boolean
check result: if same returns True, different returns False
"""
meta_list =json.loads(meta)
meta_df_user = pd.DataFrame(meta_list)
# initial check result
checkResult = True
# rename
meta_df_user.rename(columns={'name': 'variable'}, inplace=True)
# set as index
meta_df_user.set_index('variable', inplace=True)
# set roles from user input
meta_df_user['isCount'] = meta_df_user['isCount'].replace({'Y': True, 'N': False})
# set weighting_var from user input
meta_df_user['weighting_var'] = meta_df_user['weighting_var'].replace('N/A', np.nan)
# append dtype to user input metadata
meta_df_user['dtype'] = labeled_df.meta_df['dtype']
# check equal after sorting the columns
if meta_df_user.sort_index(axis=1).equals(labeled_df.meta_df.sort_index(axis=1)):
checkResult = True
else:
checkResult = False
return checkResult
def getDistanceHeatmapDict(labeled_df, cur_result_df):
"""
Generate Distance Heatmap Dictitonary List for overview
by grouping the results and extracting distances from result table.
Parameters
-----------
labeled_df : LabeledDataFrame
object from which the cur_result_df was computed, used for meta information
cur_result_df : DataFrame
A result_df or derivative of it after filtering, detecting or ranking.
Returns
--------
distance_heatmap_dict_list: Distance Heatmap Dictitonary List formatted for use in visualization
"""
distance_heatmap_dict_list = []
for trend_type, trend_df in cur_result_df.groupby(['trend_type'], sort=False):
# iterate over the GroupFeat variables
for gby, gby_trend_df in trend_df.groupby('splitby'):
# groupby the subgroups
cgby = gby_trend_df.groupby('subgroup')
# iterate over the values of the subgroups
for gby_lev,df in cgby:
distance_heatmap_dict = {}
heatmap = df.pivot(index='dependent', columns='independent', values='distance')
# replace Nan to 99
heatmap.fillna(99, inplace=True)
# trend display name
trend_display_name = labeled_df.get_trend_display_name(trend_type)
# detail view type
detail_view_type = labeled_df.get_detail_view_type(trend_type)
# overview legend type
overview_legend_type = labeled_df.get_overview_legend_type(trend_type)
distance_heatmap_dict = {'trend_type' : trend_type,
'trend_display_name': trend_display_name,
'detail_view_type': detail_view_type,
'overview_legend_type': overview_legend_type,
'splitby': gby,
'subgroup': gby_lev,
'heatmap':heatmap.to_dict('index')}
distance_heatmap_dict_list.append(distance_heatmap_dict)
return distance_heatmap_dict_list
def getOverviewLegendType(distance_heatmap_dict):
"""
Get overview legend types.
Parameters
-----------
distance_heatmap_dict : Dictionary
Distance Heatmap Dictitonary List formatted for use in visualization.
Returns
--------
overview_legend_types: List
Legend types for overview distance matrices
"""
overview_legend_types = [hd['overview_legend_type'] for hd in distance_heatmap_dict]
# get unique values in overview_legend_types
legend_types_unique = set(overview_legend_types)
overview_legend_types = list(legend_types_unique)
return overview_legend_types
def replaceTrendDisplayName(cur_result_df):
"""
Add trend display name column to df.
Parameters
-----------
cur_result_df : DataFrame
A result_df or derivative of it after filtering, detecting or ranking.
Returns
--------
cur_result_df: DataFrame
input with trend_type moved to the trend_name column and the display name in the trend_type column
"""
# compute mapping dictionary to update tend names to diplay names
name_mapper = {k:v().display_name for k,v in wg.all_trend_types.items()}
# preserve trend_type short names in the trend_name column
cur_result_df['trend_name'] = cur_result_df['trend_type']
# replace the trend_types with the display names
cur_result_df.replace({'trend_type': name_mapper}, inplace=True)
return cur_result_df
def getRankTrendDetail(labeled_df, dependent, independent, splitby):
"""
Extract stats for rank trend detail view.
Parameters
-----------
labeled_df : DataFrame
LabeledDataFrame
independent : str
a variable that will have independent information
dependent : str
a variable that will have dependent information
splitby : str
a variable that will have splitby information
Returns
--------
detail_df: dataframe
detail stats
count_df: dataframe
detail counts
"""
# trend dictionary
trend_idx_dict = {cur_trend.name: i for i, cur_trend in enumerate(labeled_df.trend_list)}
# get index for rank trend
rank_trend_idx = trend_idx_dict.get("rank_trend")
trend_precompute = labeled_df.trend_list[rank_trend_idx].trend_precompute
# aggregate' stats
sel_agg_trend = '_'.join(['rank_trend', 'agg_trend', dependent, independent])
# create a new DataFrame for detail view
detail_df = pd.DataFrame()
# create a new DataFrame for counts
count_df = | pd.DataFrame() | pandas.DataFrame |
#!/usr/bin/env python2
# -*- coding: utf-8 -*-
from __future__ import division # so that 1/3=0.333 instead of 1/3=0
from psychopy import visual, core, data, event, logging, gui
from psychopy.constants import * # things like STARTED, FINISHED
import pandas as pd
import numpy as np # whole numpy lib is available, prepend 'np.'
from numpy import sin, cos, tan, log, log10, pi, average, sqrt, std, deg2rad, rad2deg, linspace, asarray
from numpy.random import random, randint, normal, shuffle
import os # handy system and path functions
import statsmodels.formula.api as sm
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.io
# Ensure that relative paths start from the same directory as this script
_thisDir = os.path.dirname(os.path.abspath(__file__))
os.chdir(_thisDir)
# Store info about the experiment session
expName = u'r2d4_MM' # from the Builder filename that created this script
expInfo = {'participant':u'', 'session':u''}
dlg = gui.DlgFromDict(dictionary=expInfo, title=expName)
if dlg.OK == False: core.quit() # user pressed cancel
expInfo['date'] = data.getDateStr() # add a simple timestamp
expInfo['expName'] = expName
# Data file name stem = absolute path + name; later add .psyexp, .csv, .log, etc
filename = _thisDir + os.sep + 'data/%s_%s_%s' %(expInfo['participant'], expName, expInfo['date'])
out_all_fn = _thisDir + os.sep + 'data/%s_%s_%s_responses.csv' %(expInfo['participant'], expName, expInfo['session'])
data_out = pd.DataFrame(columns=('onsetTime','correctResp','keysPressed'))
# An ExperimentHandler isn't essential but helps with data saving
thisExp = data.ExperimentHandler(name=expName, version='',
extraInfo=expInfo, runtimeInfo=None,
originPath=None,
savePickle=True, saveWideText=True,
dataFileName=filename)
#save a log file for detail verbose info
logFile = logging.LogFile(filename+'.log', level=logging.EXP)
logging.console.setLevel(logging.WARNING) # this outputs to the screen, not a file
endExpNow = False # flag for 'escape' or other condition => quit the exp
# Start Code - component code to be run before the window creation
# Setup the Window
win = visual.Window(size=(500, 500), fullscr=True, screen=0, allowGUI=False, allowStencil=False,
monitor='testMonitor', color=[-1,-1,-1], colorSpace='rgb',
blendMode='avg', useFBO=True,
)
# store frame rate of monitor if we can measure it successfully
expInfo['frameRate']=win.getActualFrameRate()
if expInfo['frameRate']!=None:
frameDur = 1.0/round(expInfo['frameRate'])
else:
frameDur = 1.0/60.0 # couldn't get a reliable measure so guess
# Initialize components for Routine "Instructions"
InstructionsClock = core.Clock()
text_2 = visual.TextStim(win=win, ori=0, name='text_2',
text=u'The experiment is about to begin. ', font=u'Arial',
pos=[0, 0], height=0.1, wrapWidth=None,
color=u'white', colorSpace='rgb', opacity=1,
depth=0.0)
# Initialize components for Routine "trial"
trialClock = core.Clock()
ISI = core.StaticPeriod(win=win, screenHz=expInfo['frameRate'], name='ISI')
image = visual.ImageStim(win=win, name='image',units='pix',
image='sin', mask=None,
ori=0, pos=[0, 0], size=[200,200],
color=[1,1,1], colorSpace='rgb', opacity=1,
flipHoriz=False, flipVert=False,
texRes=128, interpolate=True, depth=0.0)
fixation = visual.ShapeStim(win,
vertices=((0, -0.075), (0, 0.075), (0,0), (-0.05,0), (0.05, 0)),
lineWidth=3,
closeShape=False,
lineColor='white')
Wrong_1 = visual.Circle(win=win, units = 'pix', radius = 100,lineColor='red', fillColor = 'red')
# Initialize components for Routine "End"
EndClock = core.Clock()
text = visual.TextStim(win=win, ori=0, name='text',
text=u'Experiment is completed. Thank you for your participation.', font=u'Arial',
pos=[0, 0], height=0.1, wrapWidth=None,
color=u'white', colorSpace='rgb', opacity=1,
depth=0.0)
#######################
#### Set up onsets ####
#######################
corr_thresh = 0.1
dfStims = pd.DataFrame
sequence_img_ids = []
img_dict = {2: 'image_folder/stim_2.png', 3: 'image_folder/stim_3.png', 4: 'image_folder/stim_4.png', 5: 'image_folder/stim_5.png'}
key_dict = {2:'2', 3:'3', 4:'4', 5:'5'}
isDone = 0
while not isDone:
trial_types = np.asarray([2, 3, 4, 5])
trial_IDs = np.asarray(range(4))
trial_freq = np.asarray([12, 12, 12, 12])
iti_range = np.asarray([2, 2, 2, 2, 3, 3, 3, 4, 5, 6, 7, 8])
n_post = 3
t_vec = []
iti_vec = []
tid_vec = []
for tt in range(0,len(trial_types)):
t_vec = np.repeat(trial_types,12)
iti_vec = np.tile(iti_range,4)
np.random.shuffle(t_vec)
np.random.shuffle(iti_vec)
vec = [0]
id_vec = vec
for t in range(0, len(t_vec)):
vec = vec + [t_vec[t]] + np.repeat(0,iti_vec[t]).tolist()
vec = vec + [0,0,0]
dfStims = pd.DataFrame()
X = np.zeros((len(vec),len(trial_types)))
ons = np.zeros((12,4))
for c in trial_types:
a = np.where(vec==c)[0]
ons[:,c-2] = a*2
for indx in range(0, len(a)):
name = a[indx]
X[a[indx]][c-2]= 1
df= | pd.DataFrame(X) | pandas.DataFrame |
import numpy as np; np.set_printoptions(precision=4, linewidth=200)
import pandas as pd; pd.set_option('display.width', 200)
import os
import logging
import scipy.stats as stats
from tqdm import tqdm
from polyfun import configure_logger, check_package_versions
from polyfun_utils import set_snpid_index
from pyarrow import ArrowIOError
from pyarrow.lib import ArrowInvalid
from polyfun_utils import DEFAULT_REGIONS_FILE
def main(args):
#read sumstats file
try:
df_sumstats = pd.read_parquet(args.sumstats)
except (ArrowIOError, ArrowInvalid):
df_sumstats = pd.read_table(args.sumstats, sep='\s+')
#compute p-values if needed
if args.pvalue_cutoff is not None:
df_sumstats['P'] = stats.chi2(1).sf(df_sumstats['Z']**2)
#read regions file
df_regions = | pd.read_table(args.regions_file) | pandas.read_table |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# @Author : Mike
# @Contact : <EMAIL>
# @Time : 2020/1/6 22:49
# @File : base.py
import lightgbm as lgb
import numpy as np
import pandas as pd
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
from scipy import sparse
from sklearn.model_selection import train_test_split
def LR_test(train_x, train_y, test_x, test_y):
from sklearn.metrics import log_loss
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(penalty='l2', dual=False, tol=0.0001, C=0.06, random_state=None,
solver='liblinear', max_iter=100,
verbose=1)
lr.fit(train_x, train_y)
predict = lr.predict_proba(test_x)[:, 1]
print(log_loss(test_y, predict))
def LGB_test(train_x, train_y, test_x, test_y, cate_col=None):
if cate_col:
data = pd.concat([train_x, test_x])
for fea in cate_col:
data[fea] = data[fea].fillna('-1')
data[fea] = LabelEncoder().fit_transform(data[fea].apply(str))
train_x = data[:len(train_x)]
test_x = data[len(train_x):]
print("LGB test")
clf = lgb.LGBMClassifier(
boosting_type='gbdt', num_leaves=31, reg_alpha=0.0, reg_lambda=1,
max_depth=-1, n_estimators=3000, objective='binary',
subsample=0.7, colsample_bytree=0.7, subsample_freq=1, # colsample_bylevel=0.7,
learning_rate=0.01, min_child_weight=25, random_state=2018, n_jobs=50
)
clf.fit(train_x, train_y, eval_set=[(train_x, train_y), (test_x, test_y)], early_stopping_rounds=100)
feature_importances = sorted(zip(train_x.columns, clf.feature_importances_), key=lambda x: x[1])
return clf.best_score_['valid_1']['binary_logloss'], feature_importances
def LGB_predict(data, file):
import math
data = data.drop(['hour48', 'hour', 'user_id', 'shop_id', 'query1', 'query',
'item_property_list', 'context_id', 'context_timestamp', 'predict_category_property'], axis=1)
data['item_category_list'] = LabelEncoder().fit_transform(data['item_category_list'])
train = data[data['is_trade'] > -1]
predict = data[data['is_trade'] == -2]
res = predict[['instance_id']]
train_y = train.pop('is_trade')
train_x = train.drop(['day', 'instance_id'], axis=1)
test_x = predict.drop(['day', 'instance_id', 'is_trade'], axis=1)
clf = lgb.LGBMClassifier(
boosting_type='gbdt', num_leaves=31, reg_alpha=0.0, reg_lambda=1,
max_depth=-1, n_estimators=3000, objective='binary',
subsample=0.7, colsample_bytree=0.7, subsample_freq=1, # colsample_bylevel=0.7,
learning_rate=0.01, min_child_weight=25, random_state=2018, n_jobs=50
)
clf.fit(train_x, train_y, eval_set=[(train_x, train_y)])
res['predicted_score'] = clf.predict_proba(test_x)[:, 1]
testb = pd.read_csv('../data/round2_ijcai_18_test_b_20180510.txt', sep=' ')[['instance_id']]
res = pd.merge(testb, res, on='instance_id', how='left')
res.to_csv('../submit/' + file + '.txt', sep=' ', index=False)
"""随机划分15%作为测试"""
def off_test_split(org, cate_col=None):
data = org[org.is_trade > -1]
data = data.drop(
['hour48', 'hour', 'user_id', 'query1', 'query',
'instance_id', 'item_property_list', 'context_id', 'context_timestamp', 'predict_category_property'], axis=1)
data['item_category_list'] = LabelEncoder().fit_transform(data['item_category_list'])
y = data.pop('is_trade')
train_x, test_x, train_y, test_y = train_test_split(data, y, test_size=0.15, random_state=2018)
train_x.drop('day', axis=1, inplace=True)
test_x.drop('day', axis=1, inplace=True)
score = LGB_test(train_x, train_y, test_x, test_y, cate_col)
return score
"""取最后2个小时作为测试"""
def off_test_2hour(org, cate_col=None):
data = org[org.is_trade > -1]
data = data.drop(
['hour48', 'user_id', 'query1', 'query',
'instance_id', 'item_property_list', 'context_id', 'context_timestamp', 'predict_category_property'], axis=1)
# data = data.drop(
# ['hour48', 'shop_score_delivery', 'shop_star_level', 'user_id', 'shop_review_num_level', 'shop_id',
# 'instance_id', 'item_property_list', 'context_id', 'context_timestamp', 'predict_category_property'], axis=1)
data['item_category_list'] = LabelEncoder().fit_transform(data['item_category_list'])
train = data[data.hour < 10]
test = data[data.hour >= 10]
train = train.drop(['hour', 'day'], axis=1)
test = test.drop(['hour', 'day'], axis=1)
train_y = train.pop('is_trade')
test_y = test.pop('is_trade')
train_x = train
test_x = test
score = LGB_test(train_x, train_y, test_x, test_y, cate_col)
return score
"""合并多部分特征,f1为train,f2为其他特征的集合"""
def add(f1, f2):
for i in f2:
f1 = | pd.merge(f1, i, on='instance_id', how='left') | pandas.merge |
import pandas as pd
import streamlit as st
import folium
from streamlit_folium import folium_static
from folium.plugins import MarkerCluster
import plotly.express as px
import geopandas
from PIL import Image
st.set_page_config(layout='wide')
@st.cache(allow_output_mutation=True)
def get_data(path):
data = pd.read_csv(path)
return data
@st.cache(allow_output_mutation=True)
def get_geofile(url):
geofile = geopandas.read_file(url)
return geofile
@st.cache(allow_output_mutation=True)
def get_image(path2):
imag1 = Image.open(path2)
return imag1
def data_groupby(data, grou):
cou = data[[grou[0], grou[1]]].groupby(grou[0]).median().reset_index()
return cou
def data_merge(data, df, mer):
merg = pd.merge(data, df, on=mer, how='inner')
return merg
def data_rename(data, cols, ren):
df = data.rename({cols[0]: ren[0], cols[1]: ren[1]}, axis='columns')
return df
def data_reset():
data = pd.read_csv('kc_house_data.csv')
return data
def data_aprimorate(data):
df2 = pd.read_csv('kc_data.csv')
return df2
def premises_plan(imag1):
st.title('1. Premises and Solution Plan:')
st.image(imag1, caption='Created by <NAME>')
st.subheader('1.1. Premises:')
st.write('This project has as premise to help the company "House Sales in King County, USA", '
'to find good home buying opportunities and then be able to resell for a higher price '
'and thus obtain a higher profit thanks to this.')
st.write('House Rocket asked on the basis of two main questions:')
st.write('1. What properties to buy and how much to sell?')
st.write('2. When is the best time to sell these properties?')
st.write('In this way, this project has as premises to be able to answer these questions, '
'and also to produce other relevant insights that may be able to have an impact in '
'decision-making and, consequently, in the business itself.')
st.subheader('1.2. Solution Plan:')
st.write('The planning can be said to be divided into 4 parts: good buying opportunities, '
'increase in the price to be able to sell, best time for resale and chances to be '
'validated or devalued. Thus, the details of each one are as follows:')
st.write('Purchasing opportunities > at this point the analysis was carried out in ascending '
'order considering the following aspects: size, price, floors, water view, number '
'of bedrooms and bathrooms. All these aspects in ascending order, that is, the more these aspects before fulfilling (eg: 2 bathrooms, 2 bedrooms, price below the median and etc ...), the better the classification of houses, considering this order of the worst even the best rating: Bad> Good> Great> Excellent')
st.write('Increase in the price of houses for resale > was carried out according to a study '
'of the average price of houses with these conditions and their offers.')
st.write('Best time for resale > was carried out according to an analysis of the average price '
'in the seasons of the year, thus finding what would be the most ideal time.')
st.write('10 hypotheses were taken, which were validated or devalued in the course of this project:')
st.write('H1 - The price of houses with conditions 3 to 4 in relation to houses with condition 1 is about 41% higher.')
st.write('H2 - The price of houses with 2 to 4 bedrooms in relation to houses without bedrooms is 38.27% higher or more.')
st.write('H3 - The price of houses with 2 to 4 bathrooms in relation to houses without a bathroom is equal to or greater than 49.30%.')
st.write('H4 - The price of houses with 2 floors in relation to houses with only 1 floor is equal to or greater than 26.10%.')
st.write('H5 - Houses with a water view have a price 67.86% higher than houses without a water view.')
st.write('H6 - The best opportunities are found mostly in south seattle.')
st.write('H7: The north has the highest priced houses.')
st.write('H8 - The annual growth in house prices is around 5%.')
st.write('H9 - The monthly house price growth is somewhat linear, always maintaining price stability.')
st.write('H10 - Houses before 1960 have a lower average price.')
return None
def data_quest(df2):
f_status = st.sidebar.multiselect('Filter status:', df2['status'].unique())
st.title('2. Business Questions:')
if (f_status != []):
df2 = df2.loc[df2['status'].isin(f_status)]
else:
df2 = df2.copy()
st.subheader('2.1. What properties to buy and how much to sell them: (Filter on the side)')
df1 = df2.copy()
st.dataframe(df1[['id', 'price', 'date', 'price_median', 'condition', 'bedrooms', 'bathrooms', 'sqft_lot',
'lot_median', 'sqft_basement',
'floors', 'status', 'exchange_upgrade']].sort_values('price', ascending=True),
height=600)
st.write('The table in question was built taking into account all the data in the database to detect the best offers and rank them accordingly as follows:')
st.write('1. bad offer: the worst offers with overvalued prices and if resold, even at the best time, it would give little profit. (about 10% more)')
st.write('2. good offer: undervalued offers that can yield a good profit due to the low purchase price. (about 20% more)')
st.write('3. great offer: subvalidas offers with more bedrooms and bathrooms that can yield a great profit. (about 30% more)')
st.write('4. excellent offer: undervalued property indications with all the benefits of others and more than one floor, can yield an excellent profit. (about 40% more)')
st.write('')
st.write('All of these increases in the resale price were calculated based on whether they were sold at the right time. (this time is indicated below)')
st.subheader('2.2. When is the best time to sell these properties:')
cols = ['seasons', 'price']
kc = data_groupby(df2, cols)
st.dataframe(kc)
st.write('The best time for sale is in the spring, as, as the table shows, at that time there is an increase in'
'the price of houses by about 4% in the other seasons.')
return None
def data_analysis(df2):
st.title('3. Insights Obtained:')
c1, c2 = st.beta_columns((1, 1))
c3, c4 = st.beta_columns((1, 1))
df2 = df2.copy()
cols = ['zipcode', 'sqft_basement']
dm1 = data_groupby(df2, cols)
df1 = data_merge(df2, dm1, 'zipcode')
cols = ['sqft_basement_x', 'sqft_basement_y']
ren = ['sqft_basement', 'basement_median']
df1 = data_rename(df1, cols, ren)
cols = ['condition', 'price']
dg1 = data_groupby(df1, cols)
#percentage1 = (dg1.iloc[0, 1] / dg1.iloc[2, 1]) - 1
#percentage2 = (dg1.iloc[0, 1] / dg1.iloc[3, 1]) - 1
#percentage = -(percentage1 + percentage2) / 2
#p1 = percentage * 100
cols = ['bedrooms', 'price']
dg2 = data_groupby(df1, cols)
#percentage1 = (dg2.iloc[0, 1] / dg2.iloc[2, 1]) - 1
#percentage2 = (dg2.iloc[0, 1] / dg2.iloc[5, 1]) - 1
#percentage1 * 100, percentage2 * 100
#percentage = -(percentage1 + percentage2) / 2
#p2 = percentage * 100
cols = ['bathrooms', 'price']
dg3 = data_groupby(df1, cols)
#percentage1 = (dg3.iloc[0, 1] / dg3.iloc[2, 1]) - 1
#percentage2 = (dg3.iloc[0, 1] / dg3.iloc[5, 1]) - 1
#percentage1 * 100, percentage2 * 100
#percentage = -(percentage1 + percentage2) / 2
#p3 = percentage * 100
cols = ['floors', 'price']
dg4 = data_groupby(df1, cols)
#percentage1 = (dg4.iloc[0, 1] / dg4.iloc[2, 1]) - 1
#p4 = -(percentage1 * 100)
cols = ['waterfront', 'price']
dg5 = data_groupby(df1, cols)
#percentage1 = (dg5.iloc[0, 1] / dg5.iloc[1, 1]) - 1
#p5 = -(percentage1 * 100)
c1.subheader('H1 - The price of houses with conditions 3 to 4 in relation to houses with '
'condition 1 is about 41% higher.(True)')
# data plot
cols = ['condition', 'price']
ren = ['Condition', 'Price Median']
dg1 = data_rename(dg1, cols, ren)
fig = px.bar(dg1.head(4), x='Condition', y='Price Median', color='Price Median', title='Median Prices Per Condition')
c1.plotly_chart(fig)
c1.write('The graph above shows that houses with conditions 3 to 4 have a price about 41% '
'higher than houses with conditions 1.')
c2.subheader('H2 - The price of houses with 2 to 4 bedrooms in relation to houses '
'without bedrooms is 38.27% higher or more. (True)')
cols = ['bedrooms', 'price']
ren = ['Bedrooms', 'Price Median']
dg2 = data_rename(dg2, cols, ren)
fig = px.bar(dg2.head(5), x='Bedrooms', y='Price Median', color='Price Median',
title='Median Prices Per Bedrooms')
c2.plotly_chart(fig)
c2.write('The graph above shows that houses with 2 to 4 bedrooms have a price of about 38.27% higher '
'than houses with no bedroom.')
c3.subheader('H3 - The price of houses with 2 to 4 bathrooms in relation to houses without '
'a bathroom is equal to or greater than 49.30%. (True)')
cols = ['bathrooms', 'price']
ren = ['Bathrooms', 'Price Median']
dg3 = data_rename(dg3, cols, ren)
fig = px.bar(dg3.head(6), x='Bathrooms', y='Price Median', color='Price Median',
title='Median Prices Per Bathrooms')
c3.plotly_chart(fig)
c3.write('The graph above shows that houses with 2 to 4 bathrooms are priced at '
'about 26.10% higher than houses with 1 without bathrooms.')
c4.subheader('H4 - The price of houses with 2 floors in relation to houses with only 1 floor '
'is equal to or greater than 26.10%. (True)')
cols = ['floors', 'price']
ren = ['Floors', 'Price Median']
dg4 = data_rename(dg4, cols, ren)
fig = px.bar(dg4.head(2), x='Floors', y='Price Median', color='Price Median',
title='Median Prices Per Floors')
c4.plotly_chart(fig)
c4.write('The graph above shows that houses with 2 floors have a price about 26.10% higher '
'than houses with 1 floor.')
st.subheader('H5 - Houses with a water view have a price 67.86% higher than houses '
'without a water view. (True)')
cols = ['waterfront', 'price']
ren = ['Waterfront', 'Price Median']
dg5 = data_rename(dg5, cols, ren)
fig = px.bar(dg5, x='Waterfront', y='Price Median', color='Price Median',
title='Median Prices Per Waterfront')
st.plotly_chart(fig)
st.write('The graph above shows that houses with a water view have a price of about 67.86% '
'higher than houses without.')
return None
def buys_map(df2, geofile):
st.title('4. Best Opportunities Vision:')
c1, c2 = st.beta_columns((1, 1))
c1.subheader('H6 - The best opportunities (excellent offers) se '
'were found on general in the south of seattle. (False)')
df = df2[df2['status'] == 'excellent offer']
# Base Map - Folium
density_map = folium.Map(location=[data['lat'].mean(), data['long'].mean()],
default_zoom_start=15)
marker_cluster = MarkerCluster().add_to(density_map)
for name, row in df.iterrows():
folium.Marker([row['lat'], row['long']],
popup='Sold R${0} on: {1}. Features: {2} sqft, {3} bedrooms, {4}'
'bathrooms, status: {5}'.format(row['price'], row['date'],
row['sqft_living'], row['bedrooms'],
row['bathrooms'], row['status'])).add_to(
marker_cluster)
with c1:
folium_static(density_map)
c1.write('This hypothesis is invalidated, as it is visible on the map that the south has fewer '
'excellent opportunities than other regions.')
# Region Price Map
c2.subheader('H7: The north has the highest priced houses. (False)')
df = df2[['price', 'zipcode']].groupby('zipcode').mean().reset_index()
df.columns = ['ZIP', 'PRICE']
df = df.sample(50)
geofile = geofile[geofile['ZIP'].isin(df['ZIP'].tolist())]
region_price_map = folium.Map(location=[df2['lat'].mean(), df2['long'].mean()],
default_zoom_start=15)
region_price_map.choropleth(data=df,
geo_data=geofile,
columns=['ZIP', 'PRICE'],
key_on='feature.properties.ZIP',
fill_color='YlOrRd',
fill_opacity=0.7,
line_opacity=0.2,
legend_name='AVG PRICE')
with c2:
folium_static(region_price_map)
c2.write('This hypothesis is invalidated, as it is visible that the higher price density '
'(higher price in the region) would be more located to the east and not to the north.')
return None
def prices_growing(df2):
st.title('5. Studies of Growth:')
c1, c2 = st.beta_columns((1, 1))
df2['date'] = pd.to_datetime(df2['date']).dt.strftime('%Y')
cols = ['date', 'price']
dg8 = data_groupby(df2, cols)
data = pd.read_csv('kc_house_data.csv')
data['date'] = | pd.to_datetime(data['date']) | pandas.to_datetime |
"""Xray object detection dataset class."""
from typing import Dict, Tuple
import cv2
import numpy as np
import os
import pandas as pd
import torch
from albumentations.core.composition import Compose
from hydra.utils import to_absolute_path
from omegaconf import DictConfig, OmegaConf
from sklearn.model_selection import train_test_split
from torch.utils.data import Dataset
from utils import load_obj, read_labels
class XrayDataset(Dataset):
"""Xray object detection dataset class."""
def __init__(
self,
dataframe: pd.DataFrame = None,
mode: str = "train",
image_dir: str = "",
cfg: DictConfig = None,
transforms: Compose = None,
):
"""
Prepare data for object detection on chest X-ray images.
Parameters
----------
dataframe : pd.DataFrame, optional
dataframe with image id and bboxes, by default None
mode : str, optional
train/val/test, by default "train"
image_dir : str, optional
path to images, by default ""
cfg : DictConfig, optional
config with parameters, by default None
transforms : Compose, optional
albumentations, by default None
"""
self.image_dir = image_dir
self.df = dataframe
self.mode = mode
self.cfg = cfg
self.image_ids = (
os.listdir(self.image_dir)
if self.df is None
else self.df["image_id"].unique()
)
self.transforms = transforms
def __getitem__(
self, idx: int
) -> Tuple[torch.tensor, Dict[str, torch.tensor], str]:
"""
Get dataset item.
Parameters
----------
idx : int
Dataset item index
Returns
-------
Tuple[Tensor, Dict[str, Tensor], str]
(image, target, image_id)
"""
image_id = self.image_ids[idx]
image = cv2.imread(f"{self.image_dir}/{image_id}", cv2.IMREAD_COLOR)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB).astype(np.float32)
# test dataset must have some values so that transforms work.
target = {
"labels": torch.as_tensor([[0]], dtype=torch.float32),
"boxes": torch.as_tensor([[0, 0, 0, 0]], dtype=torch.float32),
}
# for train and valid test create target dict.
if self.mode != "test":
image_data = self.df.loc[self.df["image_id"] == image_id]
boxes = image_data[["x", "y", "x1", "y1"]].values
boxes = torch.as_tensor(boxes, dtype=torch.float32)
areas = image_data["area"].values
areas = torch.as_tensor(areas, dtype=torch.float32)
label_dict = read_labels(
to_absolute_path(self.cfg.data.labels_path)
)
labels = [
label_dict[label] for label in image_data["label"].values
]
labels = torch.as_tensor(labels, dtype=torch.int64)
iscrowd = torch.zeros((image_data.shape[0],), dtype=torch.int64)
target["boxes"] = boxes
target["labels"] = labels
target["image_id"] = torch.tensor([idx])
target["area"] = areas
target["iscrowd"] = iscrowd
if self.transforms:
image_dict = {
"image": image,
"bboxes": target["boxes"],
"labels": labels,
}
image_dict = self.transforms(**image_dict)
image = image_dict["image"]
target["boxes"] = torch.as_tensor(
image_dict["bboxes"], dtype=torch.float32
)
else:
image_dict = {
"image": image,
"bboxes": target["boxes"],
"labels": target["labels"],
}
image = self.transforms(**image_dict)["image"]
return image, target, image_id
def __len__(self) -> int:
"""
Get dataset size.
Returns
-------
int
Dataset size
"""
return len(self.image_ids)
def get_training_dataset(cfg: DictConfig = None) -> Dict[str, Dataset]:
"""
Get training and validation datasets.
Parameters
----------
cfg : DictConfig, optional
Project configuration, by default None
Returns
-------
Dict[str, Dataset]
{"train": train_dataset, "valid": valid_dataset}
"""
images_dir = to_absolute_path(cfg.data.images_folder_path)
data = pd.read_csv(to_absolute_path(cfg.data.dataset_path))
data["x1"] = data["x"] + data["w"]
data["y1"] = data["y"] + data["h"]
data["area"] = data["w"] * data["h"]
train_ids, valid_ids = train_test_split(
data["image_id"].unique(),
test_size=cfg.data.validation_split,
random_state=cfg.training.seed,
)
# for fast training
if cfg.training.debug:
train_ids = train_ids[:10]
valid_ids = valid_ids[:10]
train_df = data.loc[data["image_id"].isin(train_ids)]
valid_df = data.loc[data["image_id"].isin(valid_ids)]
train_augs_list = [
load_obj(i["class_name"])(**i["params"])
for i in cfg["augmentation"]["train"]["augs"]
]
train_bbox_params = OmegaConf.to_container(
(cfg["augmentation"]["train"]["bbox_params"])
)
train_augs = Compose(train_augs_list, bbox_params=train_bbox_params)
valid_augs_list = [
load_obj(i["class_name"])(**i["params"])
for i in cfg["augmentation"]["valid"]["augs"]
]
valid_bbox_params = OmegaConf.to_container(
(cfg["augmentation"]["valid"]["bbox_params"])
)
valid_augs = Compose(valid_augs_list, bbox_params=valid_bbox_params)
train_dataset = XrayDataset(train_df, "train", images_dir, cfg, train_augs)
valid_dataset = XrayDataset(valid_df, "valid", images_dir, cfg, valid_augs)
return {"train": train_dataset, "valid": valid_dataset}
def get_test_dataset(cfg: DictConfig = None) -> Dataset:
"""
Get test dataset.
Parameters
----------
cfg : DictConfig, optional
Project configuration, by default None
Returns
-------
Dataset
Pytorch dataset
"""
images_dir = to_absolute_path(cfg.data.images_folder_path)
data_path = to_absolute_path(cfg.data.dataset_path)
train_ids = set(pd.read_csv(data_path).image_id.values)
all_ids = set(os.listdir(to_absolute_path(images_dir)))
test_ids = all_ids.difference(train_ids)
# for fast testing
if cfg.testing.debug:
test_ids = list(test_ids)[:100]
test_df = | pd.DataFrame(test_ids, columns=["image_id"]) | pandas.DataFrame |
import numpy as np
import pandas as pd
from scipy.stats import norm
import unittest
import networkx as nx
from context import grama as gr
from context import models
## FD stepsize
h = 1e-8
## Core function tests
##################################################
class TestModel(unittest.TestCase):
"""Test implementation of model
"""
def setUp(self):
# Default model
self.df_wrong = pd.DataFrame(data={"z": [0.0, 1.0]})
# 2D identity model with permuted df inputs
domain_2d = gr.Domain(bounds={"x0": [-1.0, +1.0], "x1": [0.0, 1.0]})
marginals = {}
marginals["x0"] = gr.MarginalNamed(
d_name="uniform", d_param={"loc": -1, "scale": 2}
)
marginals["x1"] = gr.MarginalNamed(
sign=-1, d_name="uniform", d_param={"loc": 0, "scale": 1},
)
self.model_2d = gr.Model(
functions=[
gr.Function(
lambda x: [x[0], x[1]], ["x0", "x1"], ["y0", "y1"], "test", 0
),
],
domain=domain_2d,
density=gr.Density(marginals=marginals),
)
self.df_2d = pd.DataFrame(data={"x1": [0.0], "x0": [+1.0]})
self.res_2d = self.model_2d.evaluate_df(self.df_2d)
self.df_median_in = pd.DataFrame({"x0": [0.5], "x1": [0.5]})
self.df_median_out = pd.DataFrame({"x0": [0.0], "x1": [0.5]})
self.model_3d = gr.Model(
functions=[
gr.Function(
lambda x: x[0] + x[1] + x[2], ["x", "y", "z"], ["f"], "test", 0
)
],
density=gr.Density(marginals=marginals),
)
## Timing check
self.model_slow = gr.Model(
functions=[
gr.Function(lambda x: x, ["x0"], ["y0"], "f0", 1),
gr.Function(lambda x: x, ["x0"], ["y1"], "f1", 1),
]
)
def test_prints(self):
## Invoke printpretty
self.model_3d.printpretty()
def test_timings(self):
## Default is zero
self.assertTrue(self.model_2d.runtime(1) == 0)
## Estimation accounts for both functions
self.assertTrue(np.allclose(self.model_slow.runtime(1), 2))
## Fast function has empty message
self.assertTrue(self.model_2d.runtime_message(self.df_2d) is None)
## Slow function returns string message
msg = self.model_slow.runtime_message(pd.DataFrame({"x0": [0]}))
self.assertTrue(isinstance(msg, str))
## Basic functionality with default arguments
def test_catch_input_mismatch(self):
"""Checks that proper exception is thrown if evaluate(df) passed a
DataFrame without the proper columns.
"""
self.assertRaises(ValueError, self.model_2d.evaluate_df, self.df_wrong)
def test_var_outer(self):
## Test pass-throughs
df_test = pd.DataFrame(dict(x0=[0]))
md_no_rand = gr.Model() >> gr.cp_function(fun=lambda x: x, var=1, out=1)
md_no_rand.var_outer(pd.DataFrame(), df_det="nom")
md_no_det = md_no_rand >> gr.cp_marginals(
x0={"dist": "uniform", "loc": 0, "scale": 1}
)
md_no_det.var_outer(df_test, df_det="nom")
## Test assertions
with self.assertRaises(ValueError):
self.model_3d.var_outer(self.df_2d)
with self.assertRaises(ValueError):
self.model_3d.var_outer(self.df_2d, df_det="foo")
with self.assertRaises(ValueError):
self.model_3d.var_outer(self.df_2d, df_det=self.df_2d)
def test_drop_out(self):
"""Checks that output column names are properly dropped"""
md = gr.Model() >> gr.cp_function(lambda x: x[0] + 1, var=1, out=1)
df_in = gr.df_make(x0=[0, 1, 2], y0=[0, 1, 2])
df_true = gr.df_make(x0=[0, 1, 2], y0=[1, 2, 3])
df_res = md >> gr.ev_df(df=df_in)
self.assertTrue(gr.df_equal(df_res, df_true, close=True))
## Test re-ordering issues
def test_2d_output_names(self):
"""Checks that proper output names are assigned to resulting DataFrame
"""
self.assertEqual(
set(self.model_2d.evaluate_df(self.df_2d).columns), set(self.model_2d.out)
)
def test_quantile(self):
"""Checks that model.sample_quantile() evaluates correctly.
"""
df_res = self.model_2d.density.pr2sample(self.df_median_in)
self.assertTrue(gr.df_equal(df_res, self.df_median_out))
def test_empty_functions(self):
md = gr.Model() >> gr.cp_bounds(x=[-1, +1])
with self.assertRaises(ValueError):
gr.eval_nominal(md)
def test_nominal(self):
"""Checks the implementation of nominal values"""
md = gr.Model() >> gr.cp_bounds(
x0=[-1, +1], x1=[0.1, np.Inf], x2=[-np.Inf, -0.1],
)
df_true = gr.df_make(x0=0.0, x1=+0.1, x2=-0.1)
df_res = gr.eval_nominal(md, df_det="nom", skip=True)
self.assertTrue(gr.df_equal(df_res, df_true))
## Test sample transforms
def test_transforms(self):
## Setup
df_corr = pd.DataFrame(dict(var1=["x"], var2=["y"], corr=[0.5]))
Sigma_h = np.linalg.cholesky(np.array([[1.0, 0.5], [0.5, 1.0]]))
md = (
gr.Model()
>> gr.cp_marginals(
x=dict(dist="norm", loc=0, scale=1), y=dict(dist="norm", loc=0, scale=1)
)
>> gr.cp_copula_gaussian(df_corr=df_corr)
)
## Copula and marginals have same var_rand order
self.assertTrue(list(md.density.marginals) == md.density.copula.var_rand)
## Transforms invariant
z = np.array([0, 0])
x = md.z2x(z)
zp = md.x2z(x)
self.assertTrue(np.all(z == zp))
df_z = gr.df_make(x=0.0, y=0.0)
df_x = md.norm2rand(df_z)
df_zp = md.rand2norm(df_x)
self.assertTrue(gr.df_equal(df_z, df_zp))
## Jacobian accurate
dxdz_fd = np.zeros((2, 2))
dxdz_fd[0, :] = (md.z2x(z + np.array([h, 0])) - md.z2x(z)) / h
dxdz_fd[1, :] = (md.z2x(z + np.array([0, h])) - md.z2x(z)) / h
dxdz_p = md.dxdz(z)
self.assertTrue(np.allclose(dxdz_fd, dxdz_p))
## Test DAG construction
def test_dag(self):
md = (
gr.Model("model")
>> gr.cp_function(lambda x: x, var=1, out=1)
>> gr.cp_function(lambda x: x[0] + x[1], var=["x0", "y0"], out=1)
)
G_true = nx.DiGraph()
G_true.add_edge("(var)", "f0", label="{}".format({"x0"}))
G_true.add_edge("f0", "(out)", label="{}".format({"y0"}))
G_true.add_edge("(var)", "f1", label="{}".format({"x0"}))
G_true.add_edge("f0", "f1", label="{}".format({"y0"}))
G_true.add_edge("f1", "(out)", label="{}".format({"y1"}))
nx.set_node_attributes(G_true, "model", "parent")
self.assertTrue(
nx.is_isomorphic(
md.make_dag(),
G_true,
node_match=lambda u, v: u == v,
edge_match=lambda u, v: u == v,
)
)
class TestEvalDf(unittest.TestCase):
"""Test implementation of eval_df()
"""
def setUp(self):
self.model = models.make_test()
def test_catch_no_df(self):
"""Checks that eval_df() raises when no input df is given.
"""
self.assertRaises(ValueError, gr.eval_df, self.model)
class TestMarginal(unittest.TestCase):
def setUp(self):
self.marginal_named = gr.MarginalNamed(
d_name="norm", d_param={"loc": 0, "scale": 1}
)
def test_fcn(self):
## Invoke summary
self.marginal_named.summary()
## Correct values for normal distribution
self.assertTrue(self.marginal_named.l(0.5) == norm.pdf(0.5))
self.assertTrue(self.marginal_named.p(0.5) == norm.cdf(0.5))
self.assertTrue(self.marginal_named.q(0.5) == norm.ppf(0.5))
# --------------------------------------------------
class TestDomain(unittest.TestCase):
def setUp(self):
self.domain = gr.Domain(bounds={"x": (0, 1)})
def test_blank(self):
## Test blank domain valid
gr.Domain()
## Invoke summary
self.domain.bound_summary("x")
## Invoke summary;
self.assertTrue(self.domain.bound_summary("y").find("unbounded") > -1)
# --------------------------------------------------
class TestDensity(unittest.TestCase):
def setUp(self):
self.density = gr.Density(
marginals=dict(
x=gr.MarginalNamed(d_name="uniform", d_param={"loc": -1, "scale": 2}),
y=gr.MarginalNamed(d_name="uniform", d_param={"loc": -1, "scale": 2}),
),
copula=gr.CopulaGaussian(
["x", "y"], pd.DataFrame(dict(var1=["x"], var2=["y"], corr=[0.5]))
),
)
def test_copula_warning(self):
md = gr.Model()
with self.assertRaises(ValueError):
md.density.sample()
def test_CopulaIndependence(self):
copula = gr.CopulaIndependence(var_rand=["x", "y"])
df_res = copula.sample(seed=101)
self.assertTrue(set(df_res.columns) == set(["x", "y"]))
## Transforms invariant
z = np.array([0, 0])
u = copula.z2u(z)
zp = copula.u2z(u)
self.assertTrue(np.all(z == zp))
## Jacobian accurate
dudz_fd = np.zeros((2, 2))
dudz_fd[0, :] = (copula.z2u(z + np.array([h, 0])) - copula.z2u(z)) / h
dudz_fd[1, :] = (copula.z2u(z + np.array([0, h])) - copula.z2u(z)) / h
dudz_p = copula.dudz(z)
self.assertTrue(np.allclose(dudz_fd, dudz_p))
def test_CopulaGaussian(self):
df_corr = pd.DataFrame(dict(var1=["x"], var2=["y"], corr=[0.5]))
Sigma_h = np.linalg.cholesky(np.array([[1.0, 0.5], [0.5, 1.0]]))
copula = gr.CopulaGaussian(["x", "y"], df_corr=df_corr)
df_res = copula.sample(seed=101)
self.assertTrue(np.isclose(copula.Sigma_h, Sigma_h).all)
self.assertTrue(set(df_res.columns) == set(["x", "y"]))
## Test raises
df_corr_invalid = pd.DataFrame(
dict(var1=["x", "x"], var2=["y", "z"], corr=[0, 0])
)
with self.assertRaises(ValueError):
gr.CopulaGaussian(["x", "y"], df_corr=df_corr_invalid)
## Transforms invariant
z = np.array([0, 0])
u = copula.z2u(z)
zp = copula.u2z(u)
self.assertTrue(np.all(z == zp))
## Jacobian accurate
dudz_fd = np.zeros((2, 2))
dudz_fd[0, :] = (copula.z2u(z + np.array([h, 0])) - copula.z2u(z)) / h
dudz_fd[1, :] = (copula.z2u(z + np.array([0, h])) - copula.z2u(z)) / h
dudz_p = copula.dudz(z)
self.assertTrue(np.allclose(dudz_fd, dudz_p))
def test_conversion(self):
df_pr_true = pd.DataFrame(dict(x=[0.5], y=[0.5]))
df_sp_true = pd.DataFrame(dict(x=[0.0], y=[0.0]))
df_pr_res = self.density.sample2pr(df_sp_true)
df_sp_res = self.density.pr2sample(df_pr_true)
self.assertTrue(gr.df_equal(df_pr_true, df_pr_res))
self.assertTrue(gr.df_equal(df_sp_true, df_sp_res))
def test_sampling(self):
df_sample = self.density.sample(n=1, seed=101)
self.assertTrue(set(df_sample.columns) == set(["x", "y"]))
# --------------------------------------------------
class TestFunction(unittest.TestCase):
def setUp(self):
self.fcn = gr.Function(lambda x: x, ["x"], ["x"], "test", 0)
self.fcn_vec = gr.FunctionVectorized(lambda df: df, ["x"], ["x"], "test", 0)
self.df = | pd.DataFrame({"x": [0]}) | pandas.DataFrame |
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import analyze
from utils import plot_collections, bin, modify, plotting
"""
Blue: #0C5DA5
Green: #00B945
"""
plt.style.use(['science', 'ieee', 'std-colors'])
fig, ax = plt.subplots()
size_x_inches, size_y_inches = fig.get_size_inches()
plt.close(fig)
sciblue = '#0C5DA5'
scigreen = '#00B945'
# --- --- SETUP
# --- files to read
dataset_dir = '/Users/mackenzie/Desktop/gdpyt-characterization/datasets/synthetic_overlap_noise-level1/'
base_dir = '/Users/mackenzie/Desktop/gdpyt-characterization/publication data/iteration 5/synthetic grid overlap random z nl1/'
# test coords
base_testcoords = 'test_coords/'
idpt1 = 'dx-5-60-5/test_id1_coords_static_grid-overlap-random-z-nl1.xlsx'
spct1 = 'dx-5-60-5/test_id11_coords_SPC_grid-overlap-random-z-nl1.xlsx'
idpt2 = 'dx-7.5-57.5-5/test_id2_coords_static_grid-overlap-random-z-nl1.xlsx'
spct2 = 'dx-7.5-57.5-5/test_id12_coords_SPC_grid-overlap-random-z-nl1.xlsx'
# true coords
true1 = dataset_dir + 'grid-random-z/calibration_input/calib_-15.0.txt'
true2 = dataset_dir + 'grid-random-z/test_input_dx7.5/B0000.txt'
# diameter parameters
path_diameter_params = dataset_dir + 'grid/results/calibration-SPC-spct-no_noise_cal/calib_spct_pop_defocus_stats_GridOverlapSPC_calib_nll1_SPC_no_noise_cal.xlsx'
# save ids
save_ids = ['test_id1_coords_static',
'test_id11_coords_SPC',
'test_id2_coords_static',
'test_id12_coords_SPC',
]
modifiers = [True, True, False, False]
# --- --- PERCENT DIAMETER OVERLAP
export_percent_diameter_overlap = False
plot_percent_diameter_overlap = False
if plot_percent_diameter_overlap or export_percent_diameter_overlap:
# --- read each percent diameter overlap dataframe (if available)
calculate_percent_overlap = False
for sid, modifier in zip(save_ids, modifiers):
if calculate_percent_overlap:
# --- For each test coords, calculate percent diameter overlap
for test_coord, true_coord, filt, sid in zip([idpt1, spct1, idpt2, spct2],
[true1, true1, true2, true2],
modifier,
save_ids):
dfo = analyze.calculate_particle_to_particle_spacing(
test_coords_path=base_dir + base_testcoords + test_coord,
theoretical_diameter_params_path=path_diameter_params,
mag_eff=10,
z_param='z_true',
zf_at_zero=False,
zf_param='zf_from_dia',
max_n_neighbors=5,
true_coords_path=true_coord,
maximum_allowable_diameter=55)
# filter dx 5 60 5 coords
if filt:
dfo = dfo[dfo['x'] < 820]
# save to excel
dfo.to_excel(base_dir + 'percent-overlap/{}_grid-overlap-random-z-nl1_percent_overlap.xlsx'.format(sid),
index=False)
else:
dfo = pd.read_excel(base_dir + 'percent-overlap/test_coords_percent_overlap/'
'{}_grid-overlap-random-z-nl1_percent_overlap.xlsx'.format(sid))
# --- --- EVALUATE RMSE Z
# limit percent diameter overlap to -25% (not overlapping here)
dfo['percent_dx_diameter'] = dfo['percent_dx_diameter'].where(dfo['percent_dx_diameter'] > -0.5, -0.5)
# binning
columns_to_bin = ['z_true', 'percent_dx_diameter']
bin_z = [-27.5, -15, -2.5, 10, 22.5]
bin_pdo = 3
dfbicts = analyze.evaluate_2d_bin_local_rmse_z(df=dfo,
columns_to_bin=columns_to_bin,
bins=[bin_z, bin_pdo],
round_to_decimals=[3, 4],
min_cm=0.5,
equal_bins=[False, True])
# --- --- PLOT RMSE Z
if plot_percent_diameter_overlap:
# Plot rmse z + number of particles binned as a function of percent diameter overlap for different z bins
fig, [ax, ax2] = plt.subplots(nrows=2, sharex=True, figsize=(size_x_inches*1.35, size_y_inches*1.5))
for name, df in dfbicts.items():
ax.plot(df.bin, df.rmse_z, '-o', label=name)
ax2.plot(df.bin, df.num_bind, '-o')
ax.set_ylabel(r'z r.m.s. error ($\mu m$)')
ax.set_yscale('log')
ax.legend(loc='upper left', bbox_to_anchor=(1, 1), title=r'$z_{bin}$')
ax2.set_xlabel(r'$\gamma \: $(\%)')
ax2.set_ylabel(r'$N_{p}$')
plt.tight_layout()
plt.savefig(base_dir + 'percent-overlap/{}_rmsez_num-binned_pdo.png'.format(sid))
plt.show()
# --- --- EXPORT RMSE Z TO EXCEL
# dfstack = modify.stack_dficts_by_key(dfbicts, drop_filename=False)
# dfstack.to_excel(base_dir + 'percent-overlap/{}_binned_rmsez_by_z_pdo.xlsx'.format(sid), index=False)
# --- --- PLOT OTHER METRICS
# --- calculate the local rmse_z uncertainty
num_bins = 25
bin_list = np.round(np.linspace(-1.25, 1, 10), 4)
min_cm = 0.5
z_range = [-40.1, 40.1]
round_to_decimal = 4
df_ground_truth = None
# bin by percent diameter overlap
if plot_percent_diameter_overlap:
dfob = bin.bin_local_rmse_z(df=dfo, column_to_bin='percent_dx_diameter', bins=bin_list, min_cm=min_cm, z_range=z_range,
round_to_decimal=round_to_decimal, df_ground_truth=df_ground_truth)
fig, ax = plt.subplots()
ax.plot(dfob.index, dfob.rmse_z, '-o')
ax.set_xlabel(r'$\gamma \: $(\%)')
ax.set_ylabel(r'z r.m.s. error ($\mu m$)')
plt.tight_layout()
plt.savefig(base_dir + 'percent-overlap/{}_binned_rmsez_by_pdo.png'.format(sid))
plt.show()
# bin by z
dfobz = bin.bin_local_rmse_z(df=dfo, column_to_bin='z_true', bins=num_bins, min_cm=min_cm, z_range=z_range,
round_to_decimal=round_to_decimal, df_ground_truth=df_ground_truth)
fig, ax = plt.subplots()
ax.plot(dfobz.index, dfobz.rmse_z, '-o')
ax.set_xlabel(r'$z_{true} \:$ ($\mu m$)')
ax.set_ylabel(r'z r.m.s. error ($\mu m$)')
plt.tight_layout()
plt.savefig(base_dir + 'percent-overlap/{}_binned_rmsez_by_z.png'.format(sid))
plt.show()
# --- --- CALCULATE RMSE BY PARTICLE TO PARTICLE SPACINGS
# --- setup
column_to_split = 'x'
round_x_to_decimal = 0
if modifier:
splits = np.array([93.0, 189.0, 284.0, 380.0, 475.0, 571.0, 666.0, 762.0, 858.0, 930]) # , 900.0
keys = [60, 5, 10, 15, 20, 25, 30, 35, 40, 50] # , 47.5
else:
splits = np.array([79.0, 163.5, 254.0, 348.5, 447.0, 555.5, 665.0, 777.5, 900.0])
keys = [7.5, 12.5, 17.5, 22.5, 27.5, 32.5, 37.5, 42.5, 47.5]
# --- split df into dictionary
dfsplicts_gdpyt = modify.split_df_and_merge_dficts(dfo, keys, column_to_split, splits, round_x_to_decimal)
# --- rmse z by binning x
dfmbicts_gdpyt = analyze.calculate_bin_local_rmse_z(dfsplicts_gdpyt, column_to_split, splits, min_cm, z_range,
round_x_to_decimal, dficts_ground_truth=None)
# --- plot global uncertainty - gdpyt
if plot_percent_diameter_overlap:
xlabel_for_keys = r'$\delta x (pix)$'
h = 80
scale_fig_dim = [1, 1]
fig, ax, ax2 = plotting.plot_dfbicts_global(dfmbicts_gdpyt, parameters='rmse_z', xlabel=xlabel_for_keys, h=h,
scale=scale_fig_dim)
plt.tight_layout()
plt.savefig(base_dir + 'percent-overlap/{}_global_binned_rmsez_by_z.png'.format(sid))
plt.show()
# --- --- EXPORT GLOBAL RMSE Z TO EXCEL
if export_percent_diameter_overlap:
dfstack = modify.stack_dficts_by_key(dfmbicts_gdpyt, drop_filename=False)
dfstack.to_excel(base_dir + 'percent-overlap/{}_global_binned_rmsez_by_particle_spacing.xlsx'.format(sid), index=False)
# --- --- COMBINED PARTICLE TO PARTICLE SPACING
plot_particle_spacing = False
if plot_particle_spacing:
# read files
read_dir = base_dir + 'percent-overlap/particle-to-particle-spacing/'
fn1 = 'test_id1_coords_static_global_binned_rmsez_by_particle_spacing'
fn2 = 'test_id2_coords_static_global_binned_rmsez_by_particle_spacing'
fn11 = 'test_id11_coords_SPC_global_binned_rmsez_by_particle_spacing'
fn12 = 'test_id12_coords_SPC_global_binned_rmsez_by_particle_spacing'
df1 = | pd.read_excel(read_dir + fn1 + '.xlsx') | pandas.read_excel |
from django.shortcuts import render
from django.http import HttpResponse
from datetime import datetime
import psycopg2
import math
import pandas as pd
from openpyxl import Workbook
import csv
import random
def psql_pdc(query):
#credenciales PostgreSQL produccion
connP_P = {
'host' : '10.150.1.74',
'port' : '5432',
'user':'postgres',
'password':'<PASSWORD>',
'database' : 'postgres'}
#conexion a PostgreSQL produccion
conexionP_P = psycopg2.connect(**connP_P)
#print('\nConexión con el servidor PostgreSQL produccion establecida!')
cursorP_P = conexionP_P.cursor ()
#ejecucion query telefonos PostgreSQL
cursorP_P.execute(query)
anwr = cursorP_P.fetchall()
cursorP_P.close()
conexionP_P.close()
return anwr
def to_horiz(anwr_P,name,_id):
#vertical horizontal
anwr_P1 = anwr_P.pivot(index=0,columns=1)
anwr_P1[_id] = anwr_P1.index
col1 = []
i=0
for i in range(anwr_P1.shape[1]-1):
col1.append(name+str(i+1))
col1.append(_id)
anwr_P1.columns = col1
return anwr_P1
def csv_o(fn,name):
response = HttpResponse(content_type = "text/csv")
content = "attachment; filename = %s"%name
response["Content-Disposition"] = content
# for j in range(fn.shape[1]):
# try:
# fn.iloc[:,j] = fn.iloc[:,j].str.decode(encoding='utf-8-sig')
# fn.iloc[:,j] = fn.iloc[:,j].str.encode(encoding='utf_16_le')
# except:
# pass
fn2 = [tuple(x) for x in fn.values]
writer = csv.writer(response,delimiter ='|')
writer.writerow(fn.columns)
writer.writerows(fn2)
return response
def excel(fn,name):
wb = Workbook()
ws = wb.active
k = 0
a = pd.DataFrame(fn.columns)
for k in range(a.shape[0]):
ws.cell(row = 1, column = k+1).value = a.iloc[k,0]
i=0
j=0
for i in range(fn.shape[0]):
for j in range(0,fn.shape[1]):
try:
ws.cell(row = i+2, column = j+1).value = fn.iloc[i,j]
except:
pass
response = HttpResponse(content_type = "application/ms-excel")
content = "attachment; filename = %s"%name
response["Content-Disposition"] = content
wb.save(response)
return response
def excel_CV_COL(request):
today = datetime.now()
tablename = "CV_Col"+today.strftime("%Y%m%d%H") + ".xlsx"
with open("./hello/Plantillas/Colp/QueryTel_COL.txt","r") as f1:
queryP_PT = f1.read()
with open("./hello/Plantillas/Colp/QueryCor_COL.txt","r") as f2:
queryP_PC = f2.read()
with open("./hello/Plantillas/Colp/QueryDir_COL.txt","r") as f3:
queryP_PD = f3.read()
with open("./hello/Plantillas/Colp/QueryCV_COL.txt","r") as f4:
queryP_cons = f4.read()
with open("./hello/Plantillas/Colp/QueryCiu_COL.txt","r") as f6:
queryP_Ciu = f6.read()
anwr = psql_pdc(queryP_PT)
anwrC = psql_pdc(queryP_PC)
anwrD = psql_pdc(queryP_PD)
anwrCi = psql_pdc(queryP_Ciu)
yanwr = psql_pdc(queryP_cons)
anwr_P = pd.DataFrame(anwr)
anwr_C = pd.DataFrame(anwrC)
anwr_D = pd.DataFrame(anwrD)
anwr_Ci = pd.DataFrame(anwrCi)
df = pd.DataFrame(yanwr)
inf = to_horiz(anwr_P,'phone',"deudor_id")
infC = to_horiz(anwr_C,'mail',"deudor_id")
infD = to_horiz(anwr_D,'address',"deudor_id")
infCi = to_horiz(anwr_Ci,'town',"deudor_id")
df = df.rename(columns={0:'rownumber',
1:'obligacion_id',
2:'deudor_id',
3:'unico',
4:'estado',
5:'tipo_cliente',
6:'nombre',
7:'producto',
8:'initial_bucket',
9:'ciudad',
10:'sucursal',
11:'tipo_prod',
12:'dias_mora_inicial',
13:'dias_mora_actual',
14:'rango_mora_inicial',
15:'rango_mora_final',
16:'rango',
17:'suma_pareto',
18:'rango_pareto',
19:'fcast',
20:'fdesem',
21:'vrdesem',
22:'saldo_total_inicial',
23:'saldo_total_actual',
24:'saldo_capital_inicial',
25:'saldo_capital_actual',
26:'saldo_vencido_inicial',
27:'saldo_vencido_actual',
28:'pagomin',
29:'fultpago',
30:'vrultpago',
31:'agencia',
32:'tasainter',
33:'feultref',
34:'ultcond',
35:'fasigna',
36:'eqasicampana',
37:'diferencia_pago',
38:'pago_preliminar',
39:'pago_cliente',
40:'min',
41:'tarifa',
42:'honorarios',
43:'perfil_mes_4',
44:'perfil_mes_3',
45:'perfil_mes_2',
46:'perfil_mes_1',
47:'fecha_primer_gestion',
48:'fecha_ultima_gestion',
49:'perfil_mes_actual',
50:'contactabilidad',
51:'ultimo_alo',
52:'descod1',
53:'descod2',
54:'asesor',
55:'fecha_gestion',
56:'telefono_mejor_gestion',
57:'mejorgestionhoy',
58:'asesor_indicador_hoy',
59:'repeticion',
60:'llamadas',
61:'sms',
62:'correos',
63:'gescall',
64:'visitas',
65:'whatsapp',
66:'no_contacto',
67:'total_gestiones',
68:'telefono_positivo',
69:'marcaciones_telefono_positivo',
70:'ultima_marcacion_telefono_positivo',
71:'fec_creacion_ult_compromiso',
72:'fec_pactada_ult_compromiso',
73:'valor_acordado_ult_compromiso',
74:'asesor_ult_compromiso',
75:'cantidad_acuerdos_mes',
76:'estado_acuerdo',})
fn = pd.merge(df,inf,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infC,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infD,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infCi,on = ["deudor_id"]\
,how = "left",indicator = False)
return excel(fn,tablename)
def csv_CV_Claro(request):
today = datetime.now()
tablename = "CV_Claro" + today.strftime("%Y%m%d%H") + ".csv"
with open("./hello/Plantillas/Claro/QueryTel_Claro.txt","r") as f1:
queryP_PT = f1.read()
with open("./hello/Plantillas/Claro/QueryCor_Claro.txt","r") as f2:
queryP_PC = f2.read()
with open("./hello/Plantillas/Claro/QueryCV_Claro.txt","r") as f4:
queryP_cons = f4.read()
anwr = psql_pdc(queryP_PT)
anwrC = psql_pdc(queryP_PC)
yanwr = psql_pdc(queryP_cons)
#dataframes
anwr_P = pd.DataFrame(anwr)
anwr_C = pd.DataFrame(anwrC)
df = pd.DataFrame(yanwr)
anwr_P1 = to_horiz(anwr_P,'phone','deudor_id')
#renombrar campos correos
anwr_C = anwr_C.rename(columns={
0:'deudor_id',
1:'mail0',
2:'mail1'})
anwr_C1 = anwr_C.drop_duplicates(subset=['deudor_id'])
#renombrar campos CV
df = df.rename(columns={0:'rownumber',
1:'deudor_id',
2:'obligacion_id',
3:'nombredelcliente',
4:'estado',
5:'tipo_cliente',
6:'unico',
7:'crmorigen',
8:'potencialmark',
9:'prepotencialmark',
10:'writeoffmark',
11:'dias_mora',
12:'segmento_bpo',
13:'rango_bpo',
14:'tipo',
15:'fecha_de_vencimiento',
16:'min_cliente',
17:'valorscoring',
18:'numeroreferenciadepago',
19:'monto_inicial',
20:'monto_ini_cuenta',
21:'porcentaje_descuento',
22:'valor_descuento',
23:'valor_a_pagar',
24:'deuda_real',
25:'valor_pago',
26:'saldo_pendiente',
27:'fecha_pago',
28:'fecha_compromiso',
29:'fecha_pago_compromiso',
30:'valor_compromiso',
31:'estado_acuerdo',
32:'ind_m4',
33:'ind_m3',
34:'ind_m2',
35:'ind_m1',
36:'fecha_primer_gestion',
37:'fecha_ultima_gestion',
38:'indicador',
39:'phone',
40:'asesor',
41:'fecha_gestion',
42:'contactabilidad',
43:'indicador_hoy',
44:'repeticion',
45:'llamadas',
46:'sms',
47:'correos',
48:'gescall',
49:'whatsapp',
50:'visitas',
51:'no_contacto',
52:'total_gestiones',
53:'telefono_positivo',
54:'fec_ultima_marcacion'})
#a = fn[fn.obligacion_id == '9876510000211227']
#i=0
#lin = ['no_contacto_mes_actual','gescall_mes_actual','tel_mes_actual','tel_positivo']
#for i in lin:
# df[i].fillna(0,inplace=True)
# df[i] = df[i].apply(lambda x: round(x))
# df[i] = df[i].astype('str')
fn = pd.merge(df,anwr_P1,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,anwr_C1,on = ["deudor_id"]\
,how = "left",indicator = False)
return csv_o(fn,tablename)
def csv_CV_CarP(request):
today = datetime.now()
tablename = "CV_CarP" + today.strftime("%Y%m%d%H") + ".csv"
with open("./hello/Plantillas/CarP/QueryTel_CarP.txt","r") as f1:
queryP_PT = f1.read()
with open("./hello/Plantillas/CarP/QueryCor_CarP.txt","r") as f2:
queryP_PC = f2.read()
with open("./hello/Plantillas/CarP/QueryDir_CarP.txt","r") as f3:
queryP_PD = f3.read()
with open("./hello/Plantillas/CarP/QueryCV_CarP.txt","r") as f4:
queryP_cons = f4.read()
with open("./hello/Plantillas/CarP/QueryCiu_CarP.txt","r") as f5:
queryP_Ciu = f5.read()
anwr = psql_pdc(queryP_PT)
anwrC = psql_pdc(queryP_PC)
anwrD = psql_pdc(queryP_PD)
anwrCi = psql_pdc(queryP_Ciu)
yanwr = psql_pdc(queryP_cons)
anwr_P = pd.DataFrame(anwr)
anwr_C = pd.DataFrame(anwrC)
anwr_D = pd.DataFrame(anwrD)
anwr_Ci = pd.DataFrame(anwrCi)
df = pd.DataFrame(yanwr)
inf = to_horiz(anwr_P,'phone',"deudor_id")
infC = to_horiz(anwr_C,'mail',"deudor_id")
infD = to_horiz(anwr_D,'address',"deudor_id")
infCi = to_horiz(anwr_Ci,'town',"deudor_id")
#renombrar campos CV
df = df.rename(columns={0:'deudor_id',
1:'unico',
2:'nombre',
3:'obligacion',
4:'obligacion_17',
5:'tipo_cliente',
6:'sucursal_final',
7:'zona',
8:'ano_castigo',
9:'saldo_k_pareto_mes_vigente',
10:'intereses',
11:'honorarios_20',
12:'saldo_total_mes_vigente',
13:'saldo_total_pareto_mes_vigente_',
14:'saldokpareto',
15:'rango_k_pareto',
16:'interesespareto',
17:'honorariospareto',
18:'porcentaje_k_del_total',
19:'porcentaje_intereses_del_total',
20:'porcentaje_honorarios_del_total',
21:'rango_k_porcentaje',
22:'capital_20_porciento',
23:'dias_mora_acumulado',
24:'marca_juridica_cliente',
25:'focos',
26:'valor_pago',
27:'ultima_fecha_pago',
28:'estado_cliente_mes_anterior',
29:'valor_compromiso',
30:'fecha_compromiso',
31:'fecha_pactada_compromiso',
32:'asesor_compromiso',
33:'ind_m4',
34:'ind_m3',
35:'ind_m2',
36:'ind_m1',
37:'fecha_primer_gestion',
38:'fecha_ultima_gestion',
39:'indicador',
40:'telefono_mejor_gestion',
41:'asesor_mejor_gestion',
42:'fecha_gestion',
43:'contactabilidad',
44:'indicador_hoy',
45:'repeticion',
46:'llamadas',
47:'sms',
48:'correos',
49:'gescall',
50:'whatsapp',
51:'visitas',
52:'no_contacto',
53:'total_gestiones',
54:'telefono_positivo',
55:'fec_ultima_marcacion',
56:'investigacion_de_bienes'})
fn = pd.merge(df,inf,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infC,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infD,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infCi,on = ["deudor_id"]\
,how = "left",indicator = False)
return csv_o(fn,tablename)
def csv_CV_FalaJ(request):
today = datetime.now()
tablename = "CV_FalJ"+today.strftime("%Y%m%d%H") + ".csv"
with open("./hello/Plantillas/Fala/QueryTel_Fal.txt","r") as f1:
queryP_PT = f1.read()
with open("./hello/Plantillas/Fala/QueryCor_Fal.txt","r") as f2:
queryP_PC = f2.read()
with open("./hello/Plantillas/Fala/QueryDir_Fal.txt","r") as f3:
queryP_PD = f3.read()
with open("./hello/Plantillas/Fala/QueryCV_FalJ.txt","r") as f4:
queryP_cons = f4.read()
with open("./hello/Plantillas/Fala/QueryPa_Fal.txt","r") as f5:
queryP_Pa = f5.read()
with open("./hello/Plantillas/Fala/QueryRe_Fal.txt","r") as f6:
queryP_PR = f6.read()
with open("./hello/Plantillas/Fala/QueryCiu_Fal.txt","r") as f7:
queryP_Ciu = f7.read()
anwr = psql_pdc(queryP_PT)
anwrC = psql_pdc(queryP_PC)
anwrD = psql_pdc(queryP_PD)
anwrPa = psql_pdc(queryP_Pa)
anwrR = psql_pdc(queryP_PR)
anwrCi = psql_pdc(queryP_Ciu)
yanwr = psql_pdc(queryP_cons)
anwr_P = pd.DataFrame(anwr)
anwr_C = pd.DataFrame(anwrC)
anwr_D = pd.DataFrame(anwrD)
anwr_Pa = pd.DataFrame(anwrPa)
anwr_R = pd.DataFrame(anwrR)
anwr_Ci = pd.DataFrame(anwrCi)
df = pd.DataFrame(yanwr)
inf = to_horiz(anwr_P,'phone',"deudor_id")
infC = to_horiz(anwr_C,'mail',"deudor_id")
infD = to_horiz(anwr_D,'address',"deudor_id")
infR = to_horiz(anwr_R,'referencia',"deudor_id")
infCi = to_horiz(anwr_Ci,'town',"deudor_id")
try:
infP = to_horiz(anwr_Pa,'pago','obligacion_id')
if infP.shape[1] > 4:
tipos = infP.dtypes.to_frame()
tipos['index'] = range(len(tipos))
tipos = tipos.set_index('index')
su = []
for n in range(len(tipos)):
if str(tipos.iloc[n,0]) == 'float64':
su.append(n)
else:
pass
infP1 = infP[['pago1','pago2']]
infP1['pago3'] = infP.iloc[:,2:max(su)+1].sum(axis = 1)
infP1['obligacion_id'] = infP.index
infP = infP1
i=0
lin = ['pago1','pago2','pago3']
for i in lin:
infP[i].fillna(0,inplace=True)
infP[i] = infP[i].apply(lambda x: round(x))
infP[i] = '$' + infP[i].astype('str')
except:
pass
#renombrar campos CV
df = df.rename(columns={0:'idcbpo',
1:'tipo_producto_asignacion',
2:'grupo',
3:'cartera',
4:'tipo_cliente',
5:'unico',
6:'unico_pro',
7:'obligacion_id',
8:'deudor_id',
9:'nombre',
10:'producto',
11:'saldototal',
12:'saldo_pareto',
13:'segmentacion',
14:'peso',
15:'alturamora_hoy',
16:'rango',
17:'dias_mora',
18:'vencto',
19:'indicador_mejor_gestion',
20:'total_gestiones',
21:'fecha_ultima_gestion',
22:'asesor_mejor_gestion',
23:'fecha_compromiso',
24:'fecha_pago_compromiso',
25:'valor_compromiso',
26:'asesor',
27:'estado_acuerdo',
28:'dias_mora_pagos',
29:'valor_pago',
30:'fecha_pago',
31:'pendiente',
32:'pago_total',
33:'nvo_status',
34:'status_refresque',
35:'nvo_status_refresque',
36:'dias_mora_refresque',
37:'pendiente_mas_gastos',
38:'vencida_mas_gastos',
39:'gastos_mora',
40:'gastos_cv',
41:'porcentaje_gasto',
42:'valor_a_mantener_sin_gxc',
43:'cv8',
44:'cv9',
45:'cv10',
46:'cv11',
47:'cv12',
48:'restructuracion',
49:'valor_restruc',
50:'pagominimo_actual',
51:'pagominimo_anterior',
52:'periodo_actual',
53:'periodo_anterior',
54:'cuota36',
55:'cuota48',
56:'cuota60',
57:'cuota72',
58:'proyectada_cargue',
59:'aplica_ajuste',
60:'fecha',
61:'diferencia',
62:'porcentaje_saldo_total',
63:'x',
64:'valor',
65:'porcentaje_participacion',
66:'ind_m4',
67:'ind_m3',
68:'ind_m2',
69:'ind_m1',
70:'fecha_primer_gestion',
71:'telefono_mejor_gestion',
72:'fecha_gestion',
73:'contactabilidad',
74:'indicador_hoy',
75:'repeticion',
76:'llamadas',
77:'sms',
78:'correos',
79:'gescall',
80:'whatsapp',
81:'visitas',
82:'no_contacto',
83:'telefono_positivo',
84:'fec_ultima_marcacion',
85:'lista_robinson'})
fn = pd.merge(df,inf,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infR,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infC,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infD,on = ["deudor_id"]\
,how = "left",indicator = False)
fn = pd.merge(fn,infCi,on = ["deudor_id"]\
,how = "left",indicator = False)
if 'infP' in locals():
# Cruce pagos
fn = pd.merge(fn,infP,on = ["obligacion_id"]\
,how = "left",indicator = False)
# ordenamiento
lt = fn.columns.tolist()
lt = lt[:29] + lt[(infP.shape[1]-1)*-1:] + lt[29:fn.shape[1]-(infP.shape[1]-1)]
fn = fn[lt]
return csv_o(fn,tablename)
def csv_CV_FalaC(request):
today = datetime.now()
tablename = "CV_FalC"+today.strftime("%Y%m%d%H") + ".csv"
with open("./hello/Plantillas/Fala/QueryTel_Fal.txt","r") as f1:
queryP_PT = f1.read()
with open("./hello/Plantillas/Fala/QueryCor_Fal.txt","r") as f2:
queryP_PC = f2.read()
with open("./hello/Plantillas/Fala/QueryDir_Fal.txt","r") as f3:
queryP_PD = f3.read()
with open("./hello/Plantillas/Fala/QueryCV_FalC.txt","r") as f4:
queryP_cons = f4.read()
with open("./hello/Plantillas/Fala/QueryPa_Fal.txt","r") as f5:
queryP_Pa = f5.read()
with open("./hello/Plantillas/Fala/QueryRe_Fal.txt","r") as f6:
queryP_PR = f6.read()
with open("./hello/Plantillas/Fala/QueryCiu_Fal.txt","r") as f7:
queryP_Ciu = f7.read()
anwr = psql_pdc(queryP_PT)
anwrC = psql_pdc(queryP_PC)
anwrD = psql_pdc(queryP_PD)
anwrPa = psql_pdc(queryP_Pa)
anwrR = psql_pdc(queryP_PR)
anwrCi = psql_pdc(queryP_Ciu)
yanwr = psql_pdc(queryP_cons)
anwr_P = pd.DataFrame(anwr)
anwr_C = pd.DataFrame(anwrC)
anwr_D = | pd.DataFrame(anwrD) | pandas.DataFrame |
from __future__ import print_function, division
"""
.. note::
These are the spectral modeling functions for SPLAT
"""
# imports: internal
import bz2
import copy
import glob
import gzip
import os
import requests
import shutil
import sys
import time
# imports: external
#import corner
import matplotlib; matplotlib.use('agg')
from matplotlib import cm
import matplotlib.pyplot as plt
import numpy
import pandas
from scipy import stats, signal
from scipy.integrate import trapz # for numerical integration
from scipy.interpolate import griddata, interp1d
import scipy.optimize as op
from astropy.io import ascii,fits # for reading in spreadsheet
from astropy.table import Table
from astropy.table import unique as tunique
import astropy.units as u
import astropy.constants as const
#from splat._initialize import *
import splat.triangle as triangle # will want to move this to corner
from splat.initialize import *
from splat.utilities import *
from splat.citations import shortRef
import splat.plot as splot
import splat.photometry as spphot
import splat.empirical as spemp
import splat.evolve as spev
from .core import Spectrum, classifyByIndex, compareSpectra, generateMask
# structure to store models that have been read in
MODELS_READIN = {}
#######################################################
#######################################################
################## MODEL LOADING ###################
#######################################################
#######################################################
def info(model=None):
if model == None:
model = list(SPECTRAL_MODELS.keys())
elif isinstance(model,str):
model = [model]
for m in model:
mdl = checkSpectralModelName(m)
if mdl == False: print('\nNew model named {} is currently loaded'.format(m))
print('\nModel {}:'.format(mdl))
if SPECTRAL_MODELS[mdl]['bibcode'] != '':
print('\tReference: {}'.format(shortRef(SPECTRAL_MODELS[mdl]['bibcode'])))
print('\tBibcode: {}'.format(SPECTRAL_MODELS[mdl]['bibcode']))
instr = numpy.array(list(SPECTRAL_MODELS[mdl]['instruments'].keys()))
numpy.sort(instr)
f = instr[0]
for i in instr[1:]: f=f+', {}'.format(i)
print('\tComputed for instruments {}'.format(f))
print('\tParameters:')
p = _loadModelParameters(mdl)
for m in SPECTRAL_MODEL_PARAMETERS_INORDER:
if m in list(p.keys()):
if SPECTRAL_MODEL_PARAMETERS[m]['type'] == 'continuous':
print('\t\t{}: {} {} to {} {}'.format(m,numpy.nanmin(p[m]),SPECTRAL_MODEL_PARAMETERS[m]['unit'],numpy.nanmax(p[m]),SPECTRAL_MODEL_PARAMETERS[m]['unit']))
else:
pval = numpy.array(list(set(p[m])))
numpy.sort(pval)
f = pval[0]
for i in pval[1:]: f=f+', {}'.format(i)
print('\t\t{}: {} {}'.format(m,f,SPECTRAL_MODEL_PARAMETERS[m]['unit']))
return
def addUserModels(folders=[],default_info={},verbose=True):
'''
:Purpose:
Reads in list of folders with properly processed model sets, checks them, and adds them to the SPECTRAL_MODELS global variable
:Required Inputs:
None
:Optional Inputs:
* :param folders = []: By default model folders are set in the .splat_spectral_models file;
alternately (or in addition) folders of models can be included as an input list.
* :param default_info = {}: default parameter set to use for models; superceded by 'info.txt' file if present in model folder
* :param verbose = False: provide verbose feedback
:Outputs:
None, simply adds new model sets to SPECTRAL_MODELS global variable
'''
# default information dictionary
if len(default_info.keys()) == 0:
default_info = {
'folder': '',
'name': '',
'citation': '',
'bibcode': '',
'altname': [],
'default': {'teff': 1500, 'logg': 5.0, 'z': 0.}}
# read in folders specified in .splat_spectral_models
if os.path.exists(HOME_FOLDER+'/'+EXTERNAL_SPECTRAL_MODELS_FILE):
with open(HOME_FOLDER+'/'+EXTERNAL_SPECTRAL_MODELS_FILE, 'r') as frd: x = frd.read()
folders.extend(x.split('\n'))
if '' in folders: folders.remove('')
# check and read in the new folders in the SPECTRAL_MODELS dictionary
if len(folders) > 0:
for i,f in enumerate(folders):
flag = 0
minfo = copy.deepcopy(default_info)
if minfo['folder'] == '': minfo['folder'] = f
if minfo['name'] == '': minfo['name'] = os.path.normpath(f).split('/')[-1]
subfiles = os.listdir(minfo['folder'])
# no duplicate models (for now)
if minfo['name'] in list(SPECTRAL_MODELS.keys()):
print('\nWarning: spectral model set {} already exists in SPECTRAL_MODELS library; ignoring this one'.format(minfo['name']))
flag = 1
# make sure RAW directory exists (indicates models have been processed)
if 'RAW' not in subfiles:
print('\nWarning: did not find a RAW directory in {}; please process this model set using splat.model._processModels()'.format(minfo['folder']))
flag = 1
# check for additional information file
if 'info.txt' not in subfiles:
print('\nWarning: did not find info.txt file in {}; using default values for model information'.format(minfo['folder']))
else:
# try:
f = minfo['folder']
with open(f+'/info.txt', 'r') as frd: x = frd.read()
lines = x.split('\n')
if '' in lines: lines.remove('')
lines = [x.split('\t') for x in lines]
minfo = dict(lines)
minfo['folder'] = f
for k in list(default_info.keys()):
if k not in list(minfo.keys()): minfo[k] = default_info[k]
for k in list(SPECTRAL_MODEL_PARAMETERS.keys()):
if k in list(minfo.keys()): minfo['default'][k] = minfo[k]
if 'default_'+k in list(minfo.keys()): minfo['default'][k] = minfo['default_'+k]
minfo['altnames'] = minfo['altnames'].split(',')
# except:
# print('\nWarning: problem reading info.txt file in {}; using default values for model information'.format(minfo['folder']))
if flag == 0:
if verbose == True: print('Adding {} models to SPLAT model set'.format(minfo['name']))
SPECTRAL_MODELS[minfo['name']] = copy.deepcopy(minfo)
del minfo
return
def _initializeModels(verbose=False):
'''
:Purpose:
Initializes the spectral model set by adding folders to splat.SPECTRAL_MODELS global variable
:Required Inputs:
None
:Optional Inputs:
* :param verbose = False: provide verbose feedback
:Outputs:
None
'''
# default information for a new model
# if len(default_info.keys()) == 0:
default_info = {
'instruments': {},
'name': '',
'citation': '',
'bibcode': '',
'altname': [],
'default': {'teff': 1500, 'logg': 5.0, 'z': 0.}}
# folders from which models are to be found
mfolders = [SPLAT_PATH+SPECTRAL_MODEL_FOLDER]
# specified in .splat_spectral_models
if os.path.exists(EXTERNAL_SPECTRAL_MODELS_FILE):
with open(EXTERNAL_SPECTRAL_MODELS_FILE, 'r') as frd: x = frd.read()
mfolders.extend(x.split('\n'))
if os.path.exists(HOME_FOLDER+'/'+EXTERNAL_SPECTRAL_MODELS_FILE):
with open(HOME_FOLDER+'/'+EXTERNAL_SPECTRAL_MODELS_FILE, 'r') as frd: x = frd.read()
mfolders.extend(x.split('\n'))
# specified in environmental variable SPLAT_SPECTRAL_MODELS
if os.environ.get('SPLAT_SPECTRAL_MODELS') != None:
mfolders.extend(str(os.environ['SPLAT_SPECTRAL_MODELS']).split(':'))
# check the model folders
if '' in mfolders: mfolders.remove('')
rm = []
for m in mfolders:
if os.path.exists(m) == False: rm.append(m)
if len(rm) > 0:
for m in rm: mfolders.remove(m)
if len(mfolders) == 0:
print('\nNo folders containing spectral models were found to be present')
return
mfolders = list(set(mfolders))
if verbose == True:
print('Spectral model folders:')
for m in mfolders: print('\t{}'.format(m))
# go through each model folder and check model names
for i,f in enumerate(mfolders):
mnames = os.listdir(f)
rm = []
for m in mnames:
if os.path.isdir(os.path.join(f,m))==False: rm.append(m)
if len(rm) > 0:
for m in rm: mnames.remove(m)
if len(mnames) > 0:
for nm in mnames:
fnm = os.path.join(f,nm)
instruments = os.listdir(fnm)
name = checkSpectralModelName(nm)
# new model name, add to global variable
# using info.txt data if available
if name == False:
name = nm
adddict = {'name': name}
definfo = copy.deepcopy(default_info)
if 'info.txt' in instruments:
with open(os.path.join(fnm,'info.txt'), 'r') as frd: x = frd.read()
lines = x.split('\n')
if '' in lines: lines.remove('')
lines = [x.split('\t') for x in lines]
adddict = dict(lines)
if 'altnames' in list(adddict.keys()): adddict['altnames'] = adddict['altnames'].split(',')
# for k in list(SPECTRAL_MODELS[list(SPECTRAL_MODELS.keys())[0]].keys()):
# if k not in list(adddict.keys()):
# if k in list(default_info.keys()): adddict[k] = definfo[k]
# else: adddict[k] = ''
# for k in list(default_info.keys()):
# if k not in list(minfo.keys()): minfo[k] = default_info[k]
# this sets the default values - it would be better to just grab one file and set the defaults that way
if 'default' not in list(adddict.keys()): adddict['default'] = {}
for k in list(SPECTRAL_MODEL_PARAMETERS.keys()):
if k in list(adddict.keys()): adddict['default'][k] = adddict[k]
if 'default_'+k in list(adddict.keys()): adddict['default'][k] = adddict['default_'+k]
# if k in list(adddict['default'].keys()): print(k,adddict['default'][k])
# print('\nWarning: did not find info.txt file in {}; using default values for model information'.format(minfo['folder']))
# adddict['name'] = nm
if 'name' not in list(adddict.keys()): adddict['name'] = name
if 'instruments' not in list(adddict.keys()): adddict['instruments'] = {}
if 'bibcode' not in list(adddict.keys()): adddict['bibcode'] = ''
SPECTRAL_MODELS[name] = adddict
if verbose==True: print('\nAdded a new model {} with parameters {}'.format(name,adddict))
del adddict, definfo
# go through instruments
rm = []
for m in instruments:
if os.path.isdir(os.path.join(fnm,m))==False: rm.append(m)
if len(rm) > 0:
for m in rm: instruments.remove(m)
if len(instruments) > 0:
for inst in instruments:
# make sure there are files in this folder
fnmi = os.path.join(fnm,inst)
mfiles = os.listdir(fnmi)
if len(mfiles) > 0:
instrument = checkInstrument(inst)
# unknown instrument; just add for now
if instrument == False:
instrument = (inst.replace(' ','-').replace('_','-')).upper()
if instrument not in list(SPECTRAL_MODELS[name]['instruments'].keys()):
SPECTRAL_MODELS[name]['instruments'][instrument] = fnmi
if verbose == True: print('\nAdding model {} and instrument {} from {}'.format(name,instrument,fnmi))
else:
if verbose == True: print('\nModel {} and instrument {}: ignoring {} as these already exists in {}'.format(name,instrument,fnmi,SPECTRAL_MODELS[name]['instruments'][instrument]))
return
_initializeModels()
# helper functions to read in raw models
def _readBurrows06(file):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = ascii.read(os.path.normpath(file),data_start=2)
if isinstance(data['LAMBDA(mic)'][0],str):
wave = numpy.array([float(l.replace('D','e')) for l in data['LAMBDA(mic)']])*u.micron
fnu = numpy.array([float(l.replace('D','e')) for l in data['FNU']])*(u.erg/u.s/u.cm/u.cm/u.Hz)
else:
wave = numpy.array(data['LAMBDA(mic)'])*u.micron
fnu = numpy.array(data['FNU'])*(u.erg/u.s/u.cm/u.cm/u.Hz)
wave = wave.to(DEFAULT_WAVE_UNIT)
flux = fnu.to(DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
# print(wave[50],fnu[50],flux[50])
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
def _readVeyette(file,skip=0):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = []
if file[-3:] == '.gz':
with gzip.open(os.path.normpath(file),'rt') as f:
for line in f:
data.append(line.replace('\n',''))
else:
with open(os.path.normpath(file),'rt') as f:
for line in f:
data.append(line.replace('\n',''))
if skip > 0: data = data[skip:]
wave = numpy.array([float(d.split()[0]) for d in data])*u.Angstrom
wave = wave.to(DEFAULT_WAVE_UNIT)
flux = numpy.array([10.**(float(d.split()[1])) for d in data])*u.erg/u.s/u.Angstrom/u.cm/u.cm
flux = flux.to(DEFAULT_FLUX_UNIT)
return wave, flux
def _readBtsettl08(file,expon=-8.):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = []
if file[-3:] == '.gz':
with gzip.open(os.path.normpath(file),'rt') as f:
for line in f:
if line[0] != '#': data.append(line.replace('- ','-').replace('-',' -').replace('D -','D-').replace('e -','e-'))
elif file[-4:] == '.bz2':
with bz2.open(os.path.normpath(file),'rt') as f:
for line in f:
if line[0] != '#': data.append(line.replace('- ','-').replace('-',' -').replace('D -','D-'))
else:
with open(os.path.normpath(file),'rt') as f:
for line in f:
if line[0] != '#': data.append(line)
wave = numpy.array([float((d.split()[0]).replace('D','e'))/1.e4 for d in data])*u.micron
wave = wave.to(DEFAULT_WAVE_UNIT)
flux = numpy.array([10.**(float(d.split()[1].replace('D','e'))+expon) for d in data])*u.erg/(u.s*u.Angstrom*u.cm**2)
flux = flux.to(DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
def _readAtmos(file):
try:
from netCDF4 import Dataset
except:
raise ValueError('\nYou must have the netCDF4 package installed, which is part of the Anaconda installation')
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
d = Dataset(file)
nu = d.variables['nu'][:]/u.cm
fnu = d.variables['fnu'][:]*u.erg/u.s/u.cm
wave = (1./nu).to(DEFAULT_WAVE_UNIT)
flux = (fnu*nu**2).to(DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
def _readMorley14(file):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = ascii.read(os.path.normpath(file),data_start=4)
freq = numpy.array(data['col1'])*u.Hz
wave = freq.to(DEFAULT_WAVE_UNIT,equivalencies=u.spectral())
flux = numpy.array(data['col2'])*u.erg/(u.s*u.Hz*u.cm**2)
flux = flux.to(DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
# this also reads in Morley et al. 2012 models
def _readSaumon12(file):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = ascii.read(os.path.normpath(file),data_start=2)
wave = numpy.array(data['col1'])*u.micron
wave = wave.to(DEFAULT_WAVE_UNIT)
flux = numpy.array(data['col2'])*u.erg/(u.s*u.Hz*u.cm**2)
flux = flux.to(DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
# Tremblin
def _readTremblin16(file):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = ascii.read(os.path.normpath(file),data_start=2)
nu = numpy.array(data['col1'])/u.cm
fnu = numpy.array(data['col2'])*u.erg/(u.s*u.cm)
wave = (1./nu).to(u.DEFAULT_WAVE_UNIT)
flux = (fnu*nu**2).to(splat.DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
flux = flux*((((10.*u.pc)/(0.1*u.Rsun)).to(u.m/u.m))**2) # scale to surface flux
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
# this also reads in old Drift models
def _readDrift(file):
if not os.access(file, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
data = ascii.read(os.path.normpath(file))
wave = numpy.array(data['col1'])*u.micron
wave = wave.to(DEFAULT_WAVE_UNIT)
flux = numpy.array(data['col2'])*u.erg/(u.s*u.cm**3)
flux = flux.to(DEFAULT_FLUX_UNIT,equivalencies=u.spectral_density(wave))
fluxsort = [x for (y,x) in sorted(zip(wave.value,flux.value))]
wavesort = sorted(wave.value)
return wavesort*DEFAULT_WAVE_UNIT, fluxsort*DEFAULT_FLUX_UNIT
# NOTE: THIS FUNCTION IS NO LONGER IN USE
def _modelName(modelset,instrument,param):
# check modelset name
mset = checkSpectralModelName(modelset)
if mset == False: raise ValueError('\nInvalid model name {} passed to splat.model._modelName()'.format(modelset))
# set defaults for parameters not included in param
filename = mset
for k in SPECTRAL_MODEL_PARAMETERS_INORDER:
if k in list(SPECTRAL_MODELS[mset]['default'].keys()):
if k in list(param.keys()): val = param[k]
else: val = SPECTRAL_MODELS[mset]['default'][k]
kstr = '_{}{}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],val)
if k == 'teff': kstr = '_{}{}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],int(val))
elif k == 'logg': kstr = '_{}{:.1f}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],float(val))
elif k == 'z': kstr = '_{}{:.1f}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],float(val)-0.0001)
filename=filename+kstr
if modeltype != '': filename=filename+'_{}.txt'.format(modeltype)
return filename
def _processOriginalModels(sedres=100,instruments=['SED','SPEX-PRISM'],verbose=True,skipraw=True,*args,**kwargs):
default_info = {
'instruments': {},
'name': '',
'bibcode': '',
'altnames': [],
'default': {'teff': 1500, 'logg': 5.0, 'z': 0.}}
# name of model
mset = False
if len(args) >= 1: mset = args[0]
mset = kwargs.get('set',mset)
mset = kwargs.get('modelset',mset)
mset = kwargs.get('model',mset)
# input folder
folder = './'
if len(args) >=2: folder = args[1]
folder = kwargs.get('folder',folder)
folder = kwargs.get('infolder',folder)
folder = kwargs.get('input_folder',folder)
# output folder
outputfolder = ''
if len(args) >=3: outputfolder = args[2]
outputfolder = kwargs.get('outfolder',outputfolder)
outputfolder = kwargs.get('output_folder',outputfolder)
if outputfolder == '': outputfolder = folder
# check folders
if not os.path.exists(folder):
raise ValueError('\nCould not find input folder {}'.format(folder))
if not os.path.exists(outputfolder):
os.makedirs(outputfolder)
modelset = checkSpectralModelName(mset)
# generate a new modelset key in SPECTRAL_MODELS
if modelset == False:
modelset = mset
adddict = {'name': modelset,'instruments': {},'bibcode': '','altnames': [],'default': {'teff': 1500, 'logg': 5.0, 'z': 0.}}
files = os.listdir(folder)
if 'info.txt' in files:
with open(os.path.join(folder,'info.txt'), 'r') as frd: x = frd.read()
lines = x.split('\n')
if '' in lines: lines.remove('')
lines = [x.split('\t') for x in lines]
adddict = dict(lines)
if 'altnames' in list(adddict.keys()): adddict['altnames'] = adddict['altnames'].split(',')
for k in list(SPECTRAL_MODELS[list(SPECTRAL_MODELS.keys())[0]].keys()):
if k not in list(adddict.keys()):
if k in list(default_info.keys()): adddict[k] = definfo[k]
else: adddict[k] = ''
for k in list(SPECTRAL_MODEL_PARAMETERS.keys()):
if k in list(adddict.keys()): adddict['default'][k] = adddict[k]
if 'default_'+k in list(adddict.keys()): adddict['default'][k] = adddict['default_'+k]
files = os.listdir(outputfolder)
if 'info.txt' not in files:
shutil.copy(os.path.join(folder,'info.txt'),os.path.join(outputfolder,'info.txt'))
SPECTRAL_MODELS[modelset] = adddict
if verbose==True: print('\nAdded a new model {} with parameters {}'.format(modelset,adddict))
del adddict
# special 'ORIGINAL' folder
files = os.listdir(folder)
if 'ORIGINAL' in files: folder = os.path.join(folder,'ORIGINAL')
# only do instruments if RAW models already exist
skipraw = skipraw and os.path.exists(outputfolder+'/RAW/')
try:
skipraw = skipraw and len(os.listdir(outputfolder+'/RAW/')) > 0
except:
skipraw = skipraw and False
if skipraw == False:
# presets for various models - these are based on how they are downloaded
if 'burrows' in modelset.lower():
readfxn = _readBurrows06
files = glob.glob(os.path.join(folder,'*.txt'))
mparam = {}
mparam['teff'] = [float(f.split('_')[0].split('T')[-1]) for f in files]
mparam['logg'] = [float(f.split('_')[1][1:]) for f in files]
mparam['z'] = [numpy.round(numpy.log10(float(f.split('_')[-1][:3]))*10.)/10. for f in files]
mparam['cld'] = [f.split('_')[2].replace('cf','nc') for f in files]
elif 'madhu' in modelset.lower():
readfxn = _readBurrows06
files = glob.glob(os.path.join(folder,'*'))
mparam = {}
mparam['teff'] = [float(f.split('_')[1].split('t')[-1]) for f in files]
mparam['logg'] = [float(f.split('_')[2].split('g')[-1]) for f in files]
mparam['z'] = [numpy.round(numpy.log10(float(f.split('_')[3].split('z')[-1]))*10.)/10. for f in files]
mparam['fsed'] = [f.split('_')[-1].lower() for f in files]
mparam['cld'] = [(f.split('/')[-1]).split('_')[0].lower() for f in files]
mparam['kzz'] = [f.split('_')[-2].lower() for f in files]
elif 'atmos' in modelset.lower():
readfxn = _readAtmos
files = glob.glob(os.path.join(folder,'*.ncdf'))
mparam = {}
mparam['teff'] = [float((f.split('/')[-1]).split('_')[1][1:]) for f in files]
mparam['logg'] = [float((f.split('/')[-1]).split('_')[2][2:]) for f in files]
mparam['z'] = [0. for f in files]
mparam['ad'] = [float((f.split('/')[-1]).split('_')[3][1:]) for f in files]
mparam['logpmin'] = [float((f.split('/')[-1]).split('_')[4][2:]) for f in files]
mparam['logpmax'] = [float((f.split('/')[-1]).split('_')[5][2:]) for f in files]
mparam['kzz'] = [(f.split('/')[-1]).split('_')[6][3:] for f in files]
mparam['broad'] = [(f.split('/')[-1]).split('_')[7] for f in files]
mparam['cld'] = [(f.split('/')[-1]).split('_')[8] for f in files]
elif modelset == 'btsettl08':
readfxn = _readBtsettl08
files = glob.glob(os.path.join(folder,'*spec.7.gz'))
mparam = {}
mparam['teff'] = [float((f.split('/'))[-1][3:6])*100. for f in files]
mparam['logg'] = [float((f.split('/'))[-1][7:10]) for f in files]
mparam['z'] = [float((f.split('/'))[-1][10:14]) for f in files]
mparam['enrich'] = [float(((f.split('/'))[-1].split('a+'))[-1][0:3]) for f in files]
elif modelset == 'nextgen99':
readfxn = _readBtsettl08
files = glob.glob(os.path.join(folder,'lte*.gz'))
mparam = {}
mparam['teff'] = [float((f.split('/'))[-1][3:6])*100. for f in files]
mparam['logg'] = [float((f.split('/'))[-1][7:10]) for f in files]
mparam['z'] = [float((f.split('/'))[-1][10:14]) for f in files]
elif modelset == 'btnextgen' or modelset == 'btcond' or modelset == 'btdusty':
readfxn = _readBtsettl08
files = glob.glob(os.path.join(folder,'lte*.bz2'))
mparam = {}
mparam['teff'] = [float((f.split('/'))[-1][3:6])*100. for f in files]
mparam['logg'] = [float((f.split('/'))[-1][7:10]) for f in files]
mparam['z'] = [float((f.split('/'))[-1][10:14]) for f in files]
if 'enrich' in list(SPECTRAL_MODELS[modelset]['default'].keys()):
mparam['enrich'] = [float(((f.split('/'))[-1].split('a+'))[-1][0:3]) for f in files]
elif modelset == 'cond01' or modelset == 'dusty01':
readfxn = _readBtsettl08
files = glob.glob(os.path.join(folder,'*7.gz'))
mparam = {}
mparam['teff'] = [float((f.split('/'))[-1][3:5])*100. for f in files]
mparam['logg'] = [float((f.split('/'))[-1][6:9]) for f in files]
mparam['z'] = [float((f.split('/'))[-1][10:13]) for f in files]
elif modelset == 'btsettl15':
readfxn = _readBtsettl08
files = glob.glob(os.path.join(folder,'*spec.7.gz'))
mparam = {}
mparam['teff'] = [float((f.split('/'))[-1][3:8])*100. for f in files]
mparam['logg'] = [float((f.split('/'))[-1][9:12]) for f in files]
mparam['z'] = [float((f.split('/'))[-1][12:16]) for f in files]
# Morley et al. 2012: no metallicity, fsed, kzz or clouds
elif modelset == 'morley12':
readfxn = _readSaumon12
files = glob.glob(os.path.join(folder,'sp*'))
mparam = {}
mparam['teff'] = [float((((f.split('/'))[-1].split('_'))[-2].split('g'))[0][1:]) for f in files]
mparam['logg'] = [2.+numpy.log10(float((((((f.split('/'))[-1].split('_'))[-2].split('g'))[1]).split('f'))[0])) for f in files]
mparam['z'] = [SPECTRAL_MODELS[modelset]['default']['z'] for f in files]
mparam['fsed'] = ['f'+((((f.split('/'))[-1].split('_'))[-2].split('g'))[1]).split('f')[-1] for f in files]
# Morley et al. 2014: no metallicity, fsed, kzz or clouds
elif modelset == 'morley14':
readfxn = _readMorley14
files = glob.glob(os.path.join(folder,'sp*'))
mparam = {}
mparam['teff'] = [float((((f.split('/'))[-1].split('_'))[-2].split('g'))[0][1:]) for f in files]
mparam['logg'] = [2.+numpy.log10(float((((((f.split('/'))[-1].split('_'))[-2].split('g'))[1]).split('f'))[0])) for f in files]
mparam['z'] = [SPECTRAL_MODELS[modelset]['default']['z'] for f in files]
mparam['fsed'] = ['f'+(((((f.split('/'))[-1].split('_'))[-2].split('g'))[1]).split('f')[-1]).split('h')[0] for f in files]
mparam['cld'] = ['h'+(((((f.split('/'))[-1].split('_'))[-2].split('g'))[1]).split('f')[-1]).split('h')[-1] for f in files]
# Saumon & Marley 2012: no metallicity, fsed, kzz or clouds
elif modelset == 'saumon12':
readfxn = _readSaumon12
files = glob.glob(os.path.join(folder,'sp*'))
mparam = {}
mparam['teff'] = [float((((f.split('/'))[-1].split('_'))[-1].split('g'))[0][1:]) for f in files]
mparam['logg'] = [2.+numpy.log10(float((((f.split('/'))[-1].split('_'))[-1].split('g'))[1].split('nc')[0])) for f in files]
mparam['z'] = [SPECTRAL_MODELS[modelset]['default']['z'] for f in files]
elif modelset == 'drift':
readfxn = _readBtsettl08
files = glob.glob(os.path.join(folder,'lte_*'))
mparam['teff'] = [float((f.split('/')[-1]).split('_')[1]) for f in files]
mparam['logg'] = [float((f.split('/')[-1]).split('_')[2][:3]) for f in files]
mparam['z'] = [float((f.split('/')[-1]).split('_')[2][3:7]) for f in files]
mparam = {}
elif modelset == 'tremblin16':
readfxn = _readTremblin16
files = glob.glob(os.path.join(folder,'*.dat'))
mparam = {}
mparam['teff'] = [float((f.split('/')[-1]).split('_')[1][1:]) for f in files]
mparam['logg'] = [float((f.split('/')[-1]).split('_')[2][1:]) for f in files]
mparam['z'] = [SPECTRAL_MODELS[modelset]['default']['z'] for f in files]
mparam['kzz'] = [float((f.split('/')[-1]).split('_')[3][1:]) for f in files]
mparam['ad'] = [float((f.split('/')[-1]).split('_')[5][1:5]) for f in files]
elif 'veyette' in modelset.lower():
readfxn = _readVeyette
files = glob.glob(os.path.join(folder,'*.gz'))
mparam = {}
mparam['teff'] = [float((f.replace('.BT-Settl','').replace('.txt.gz','').split('/')[-1]).split('_')[0][3:]) for f in files]
mparam['logg'] = [float((f.replace('.BT-Settl','').replace('.txt.gz','').split('/')[-1]).split('_')[1]) for f in files]
mparam['z'] = [float((f.replace('.BT-Settl','').replace('.txt.gz','').split('/')[-1]).split('_')[2][1:].replace('+','')) for f in files]
mparam['enrich'] = [float((f.replace('.BT-Settl','').replace('.txt.gz','').split('/')[-1]).split('_')[3][1:].replace('+','')) for f in files]
mparam['carbon'] = [float((f.replace('.BT-Settl','').replace('.txt.gz','').split('/')[-1]).split('_')[4][1:].replace('+','')) for f in files]
mparam['oxygen'] = [float((f.replace('.BT-Settl','').replace('.txt.gz','').split('/')[-1]).split('_')[5][1:].replace('+','')) for f in files]
else:
raise ValueError('\nHave not yet gotten model set {} into _processModels'.format(modelset))
if len(files) == 0:
raise ValueError('Could not find spectral model files in {}'.format(folder))
# create folders if they don't exist
if not os.path.exists(outputfolder+'/RAW/'):
os.makedirs(outputfolder+'/RAW/')
# generate photometry - skipping for now since this takes a while
# if kwargs.get('make_photometry',False) == True:
# phot_data = {}
# for p in SPECTRAL_MODEL_PARAMETERS_INORDER: phot_data[p] = []
# for f in list(FILTERS.keys()): phot_data[f] = []
# read in files
if verbose == True: print('\nIntegrating {} models into SPLAT'.format(modelset))
for i,f in enumerate(files):
try:
wv,flx = readfxn(f)
# spmodel = Spectrum(wave=wv,flux=flx)
except:
print('\nError reading in file {}; skipping'.format(f))
else:
if verbose == True:
line = ''
for k in list(mparam.keys()): line=line+'{}: {}, '.format(k,mparam[k][i])
print('Processing {}for model {}'.format(line,modelset))
# generate raw model
outputfile = outputfolder+'/RAW/'+modelset
for k in SPECTRAL_MODEL_PARAMETERS_INORDER:
if k in list(mparam.keys()):
if SPECTRAL_MODEL_PARAMETERS[k]['type'] == 'continuous':
kstr = '_{}{:.2f}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],float(mparam[k][i]))
else:
kstr = '_{}{}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],mparam[k][i])
if k == 'teff': kstr = '_{}{}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],int(mparam[k][i]))
elif k == 'z': kstr = '_{}{:.2f}'.format(SPECTRAL_MODEL_PARAMETERS[k]['prefix'],mparam[k][i]-0.0001)
outputfile=outputfile+kstr
outputfile=outputfile+'_RAW.txt'
# old way - make table and output it
t = Table([wv,flx],names=['#wavelength','surface_flux'])
t.write(outputfile,format='ascii.tab')
# now gzip it
with open(outputfile, 'rb') as f_in, gzip.open(outputfile+'.gz', 'wb') as f_out:
shutil.copyfileobj(f_in, f_out)
os.remove(outputfile)
# if successful, add 'RAW' to instruments list in SPECTRAL_MODELS global variable
SPECTRAL_MODELS[modelset]['instruments']['RAW'] = outputfolder+'/RAW/'
# generate SED model
# if make_sed == True:
# noutputfile = outputfile.replace('RAW','SED')
# # first smooth relevant piece of spectrum
# # interpret onto observed wavelength grid
# npix = numpy.floor(numpy.log(numpy.nanmax(wv.value)/numpy.nanmin(wv.value))/numpy.log(1.+1./sedres))
# wvref = [numpy.nanmin(wv.value)*(1.+1./sedres)**i for i in numpy.arange(npix)]
# # either smooth and remap if SED is higher res than original data,
# # or integral resample if original data is higher resolution than SED
# if len(wv) <= len(wvref):
# flxsm = _smooth2resolution(wv.value,flx.value,sedres)
# f = interp1d(wv.value,flxsm,bounds_error=False,fill_value=0.)
# flxout = f(wvref)
# else:
# flxout = integralResample(wv.value,flx.value,wvref)
# t = Table([wvref,flxout],names=['#wavelength','surface_flux'])
# t.write(noutputfile,format='ascii.tab')
# generate instruments
if len(instruments) > 0:
wv = getModel(set=modelset,instrument='RAW').wave
for inst in instruments:
ins = checkInstrument(inst)
if ins != False: inst = ins
# if not os.path.exists(outputfolder+'/{}/'.format(inst)): os.makedirs(outputfolder+'/{}/'.format(inst))
if verbose == True: print('Processing models for instrument {}'.format(inst))
if inst=='SPEX-PRISM':
spref = Spectrum(10001)
processModelsToInstrument(modelset=modelset,wave=spref.wave,instrument=inst)
elif inst=='SED':
INSTRUMENTS['SED']['wave_range'] = [numpy.nanmin(wv),numpy.nanmax(wv)]
INSTRUMENTS['SED']['resolution'] = sedres
processModelsToInstrument(modelset=modelset,instrument=inst)
elif ins != False:
processModelsToInstrument(modelset=modelset,instrument=inst)
else:
print('\nDo not have enough information to generate model set for instrument {}; run this separate with processModelsToInstrument()'.format(inst))
return
def processModelsToInstrument(instrument_parameters={},wunit=DEFAULT_WAVE_UNIT,funit=DEFAULT_FLUX_UNIT,pixel_resolution=4.,wave=[],wave_range=[],resolution=None,template=None,verbose=False,overwrite=False,*args,**kwargs):
'''
:Purpose:
Converts raw spectral models into instrument-specific model sets, based on pre-defined or
supplied information on wavelength range and resolution or a template spectrum
:Required Inputs:
* `modelset` or `set`: name of the model set to convert, (for now) must be included in SPLAT distribution; may also be passed as a first argument
* `instrument` or `instr`: name of the instrument to convert, either a predefined one (splat.INSTRUMENTS.keys()) or place holder for user-specified parameters; may also be passed as a second argument
:Optional Inputs:
If a predefined instrument is not used, user must supply one of the following combinations either as keywords or in an `instrument_parameters` dictionary parameter:
* `wave`: an array containing the wavelengths to sample to; resolution is assumed 2 pixels per resolution element
* `wave_range` and `resolution`: the first is a two-element array (assumed in microns if not specified), the second the effective resolution, assuming 2 pixels per resolution element
* `wunit`: the unit for the wavelength axis
* `funit`: the unit for the flux density axis
* `template`: a template spectrum object, from which the `wave` array is selected
* `pixel_resolution` = 4: the number of pixels per resolution element
* `oversample` = 5: by what factor to oversample the spectral data when smoothing
* `overscan` = 0.05: percentage of total wavelength range to overextend in order to deal with edge effects in smoothing
* `method` = 'hanning': filter design for smoothing
:Outputs:
If necessary, creates a folder in the splat.SPECTRAL_MODEL_FOLDER/[modelset]/[instrument] and outputs the model files
'''
method = kwargs.get('method','hamming')
oversample = kwargs.get('oversample',5.)
overscan = kwargs.get('overscan',0.05)
# model set
modelset = False
if len(args) >= 1: modelset = args[0]
modelset = kwargs.get('modelset',modelset)
modelset = kwargs.get('model',modelset)
modelset = kwargs.get('set',modelset)
mset = checkSpectralModelName(modelset)
if mset == False:
raise ValueError('\nInvalid model set {}'.format(modelset))
# instrument
instrument = 'SPEX-PRISM'
if len(args) >= 2: instrument = args[1]
instrument = kwargs.get('instrument',instrument)
instrument = kwargs.get('instr',instrument)
instr = checkInstrument(instrument)
# set up parameters for making model
if instr != False:
for r in ['resolution','wave_range','wunit','funit']:
instrument_parameters[r] = INSTRUMENTS[instr][r]
else:
instr = instrument.upper()
instr = instr.replace(' ','-')
# check if instrument is already set up for this model
if instr in list(SPECTRAL_MODELS[mset]['instruments'].keys()) and overwrite == False:
print('\nInstrument {} is already computed for modelset {}; set overwrite = True to overwrite these'.format(instr,mset))
return
# use a template
if isinstance(template,splat.core.Spectrum):
instrument_parameters['wave'] = template.wave
instrument_parameters['wunit'] = template.wave.unit
instrument_parameters['funit'] = template.flux.unit
instrument_parameters['wave_range'] = [numpy.nanmin(template.wave.value),numpy.nanmax(template.wave.value)]
# set wavelength unit
if 'wunit' not in list(instrument_parameters.keys()): instrument_parameters['wunit'] = wunit
if not isUnit(instrument_parameters['wunit']):
if verbose == True: print('\nWarning: could not interpet unit {} which is type {}; setting wavelength unit to {}'.format(instrument_parameters['wunit'],type(instrument_parameters['wunit'],DEFAULT_WAVE_UNIT)))
instrument_parameters['wunit'] = DEFAULT_WAVE_UNIT
# set wavelength unit
if 'funit' not in list(instrument_parameters.keys()): instrument_parameters['funit'] = funit
if not isUnit(instrument_parameters['funit']):
instrument_parameters['funit'] = DEFAULT_FLUX_UNIT
# set wave scale
if 'wave' not in list(instrument_parameters.keys()): instrument_parameters['wave'] = wave
if len(instrument_parameters['wave']) > 1:
if isUnit(instrument_parameters['wave']):
instrument_parameters['wave'] = instrument_parameters['wave'].to(instrument_parameters['wunit']).value
if isUnit(instrument_parameters['wave'][0]):
instrument_parameters['wave'] = [w.to(instrument_parameters['wunit']).value for w in instrument_parameters['wave']]
instrument_parameters['wave_range'] = [numpy.nanmin(instrument_parameters['wave']),numpy.nanmax(instrument_parameters['wave'])]
# set wavelength range
if 'wave_range' not in list(instrument_parameters.keys()):
instrument_parameters['wave_range'] = wave_range
if len(instrument_parameters['wave_range']) > 1:
if isUnit(instrument_parameters['wave_range']):
instrument_parameters['wave_range'] = instrument_parameters['wave_range'].to(instrument_parameters['wunit']).value
if isUnit(instrument_parameters['wave_range'][0]):
instrument_parameters['wave_range'] = [w.to(instrument_parameters['wunit']).value for w in instrument_parameters['wave_range']]
# set resolution
if 'resolution' not in list(instrument_parameters.keys()):
instrument_parameters['resolution'] = resolution
# generate wavelength vector if just range and resolution given
if len(instrument_parameters['wave']) <= 1 and instrument_parameters['resolution'] != None and len(instrument_parameters['wave_range']) >= 2:
effres = instrument_parameters['resolution']*pixel_resolution
npix = numpy.floor(numpy.log(numpy.nanmax(instrument_parameters['wave_range'])/numpy.nanmin(instrument_parameters['wave_range']))/numpy.log(1.+1./effres))
# print(instr,npix)
instrument_parameters['wave'] = [numpy.nanmin(instrument_parameters['wave_range'])*(1.+1./effres)**i for i in numpy.arange(npix)]
# final error check
if len(instrument_parameters['wave']) <= 1:
raise ValueError('\nCould not set up instrument parameters {}'.format(instrument_parameters))
# generate smoothing wavelength vector
a = numpy.linspace(0.,len(instrument_parameters['wave'])-1,len(instrument_parameters['wave']))
b = numpy.linspace(0.,len(instrument_parameters['wave'])-1.,oversample*len(instrument_parameters['wave']))
f = interp1d(a,instrument_parameters['wave'])
wave_oversample = f(b)
# grab the raw files
inputfolder = kwargs.get('inputfolder',os.path.normpath(SPECTRAL_MODELS[mset]['instruments']['RAW']))
files = glob.glob(os.path.normpath(inputfolder+'/*.txt'))
if len(files) == 0:
files = glob.glob(os.path.normpath(inputfolder+'/*.gz'))
if len(files) == 0:
raise ValueError('\nCould not find model files in {}'.format(inputfolder))
# set and create folder if it don't exist
outputfolder = kwargs.get('outputfolder',inputfolder.replace('RAW',instr))
# if os.path.exists(outputfolder) == True and overwrite==False:
# raise ValueError('\nModel output folder {} already exists; set overwrite=True to overwrite'.format(outputfolder))
if not os.path.exists(outputfolder):
try:
os.makedirs(outputfolder)
except:
raise OSError('\nCould not create output folder {}'.format(outputfolder))
if verbose == True: print('Processing model set {} to instrument {}'.format(mset,instr))
for i,f in enumerate(files):
if verbose == True: print('{}: Processing model {}'.format(i,f))
noutputfile = f.replace('RAW',instr).replace('.gz','')
if not os.path.exists(noutputfile) or (os.path.exists(noutputfile) and overwrite==True):
# read in the model
spmodel = Spectrum(f,ismodel=True)
# NOTE THAT THE FOLLOWING COULD BE REPLACED BY spmodel.toInstrument()
spmodel.toWaveUnit(instrument_parameters['wunit'])
spmodel.toFluxUnit(instrument_parameters['funit'])
# trim relevant piece of spectrum
dw = overscan*(numpy.nanmax(instrument_parameters['wave'])-numpy.nanmin(instrument_parameters['wave']))
wrng = [numpy.nanmax([numpy.nanmin(instrument_parameters['wave']-dw),numpy.nanmin(spmodel.wave.value)])*instrument_parameters['wunit'],\
numpy.nanmin([numpy.nanmax(instrument_parameters['wave']+dw),numpy.nanmax(spmodel.wave.value)])*instrument_parameters['wunit']]
spmodel.trim(wrng)
# print(instrument_parameters['wave'])
# map onto oversampled grid and smooth; if model is lower resolution, interpolate; otherwise integrate & resample
if len(spmodel.wave) <= len(wave_oversample):
fflux = interp1d(spmodel.wave.value,spmodel.flux.value,bounds_error=False,fill_value=0.)
flux_oversample = fflux(wave_oversample)
else:
flux_oversample = integralResample(spmodel.wave.value,spmodel.flux.value,wave_oversample)
spmodel.wave = wave_oversample*instrument_parameters['wunit']
spmodel.flux = flux_oversample*spmodel.funit
spmodel.noise = [numpy.nan for x in spmodel.wave]*spmodel.funit
spmodel.variance = [numpy.nan for x in spmodel.wave]*(spmodel.funit**2)
# smooth this in pixel space including oversample
spmodel._smoothToSlitPixelWidth(pixel_resolution*oversample,method=method)
# resample down to final wavelength scale
fluxsm = integralResample(spmodel.wave.value,spmodel.flux.value,instrument_parameters['wave'])
# output
t = Table([instrument_parameters['wave'],fluxsm],names=['#wavelength ({})'.format(spmodel.wave.unit),'surface_flux ({})'.format(spmodel.flux.unit)])
t.write(noutputfile,format='ascii.tab')
else:
if verbose == True: print('\tfile {} already exists; skipping'.format(noutputfile))
# if successful, add this instrument to SPECTRAL_MODELS global variable
SPECTRAL_MODELS[mset]['instruments'][instr] = outputfolder
return
def loadOriginalModel(model='btsettl08',instrument='UNKNOWN',file='',**kwargs):
'''
:Purpose:
Loads up an original model spectrum at full resolution/spectral range, based on filename or model parameters.
:Required Inputs:
None
:Optional Inputs:
:param: **model**: The model set to use; may be one of the following:
- *btsettl08*: (default) model set from `Allard et al. (2012) <http://adsabs.harvard.edu/abs/2012RSPTA.370.2765A>`_ with effective temperatures of 400 to 2900 K (steps of 100 K); surface gravities of 3.5 to 5.5 in units of cm/s^2 (steps of 0.5 dex); and metallicity of -3.0, -2.5, -2.0, -1.5, -1.0, -0.5, 0.0, 0.3, and 0.5 for temperatures greater than 2000 K only; cloud opacity is fixed in this model, and equilibrium chemistry is assumed. Note that this grid is not completely filled and some gaps have been interpolated (alternate designations: `btsettled`, `btsettl`, `allard`, `allard12`)
- *burrows06*: model set from `Burrows et al. (2006) <http://adsabs.harvard.edu/abs/2006ApJ...640.1063B>`_ with effective temperatures of 700 to 2000 K (steps of 50 K); surface gravities of 4.5 to 5.5 in units of cm/s^2 (steps of 0.1 dex); metallicity of -0.5, 0.0 and 0.5; and either no clouds or grain size 100 microns (fsed = 'nc' or 'f100'). equilibrium chemistry is assumed. Note that this grid is not completely filled and some gaps have been interpolated (alternate designations: `burrows`, `burrows2006`)
- *morley12*: model set from `Morley et al. (2012) <http://adsabs.harvard.edu/abs/2012ApJ...756..172M>`_ with effective temperatures of 400 to 1300 K (steps of 50 K); surface gravities of 4.0 to 5.5 in units of cm/s^2 (steps of 0.5 dex); and sedimentation efficiency (fsed) of 2, 3, 4 or 5; metallicity is fixed to solar, equilibrium chemistry is assumed, and there are no clouds associated with this model (alternate designations: `morley2012`)
- *morley14*: model set from `Morley et al. (2014) <http://adsabs.harvard.edu/abs/2014ApJ...787...78M>`_ with effective temperatures of 200 to 450 K (steps of 25 K) and surface gravities of 3.0 to 5.0 in units of cm/s^2 (steps of 0.5 dex); metallicity is fixed to solar, equilibrium chemistry is assumed, sedimentation efficiency is fixed at fsed = 5, and cloud coverage fixed at 50% (alternate designations: `morley2014`)
- *saumon12*: model set from `Saumon et al. (2012) <http://adsabs.harvard.edu/abs/2012ApJ...750...74S>`_ with effective temperatures of 400 to 1500 K (steps of 50 K); and surface gravities of 3.0 to 5.5 in units of cm/s^2 (steps of 0.5 dex); metallicity is fixed to solar, equilibrium chemistry is assumed, and no clouds are associated with these models (alternate designations: `saumon`, `saumon2012`)
- *drift*: model set from `Witte et al. (2011) <http://adsabs.harvard.edu/abs/2011A%26A...529A..44W>`_ with effective temperatures of 1700 to 3000 K (steps of 50 K); surface gravities of 5.0 and 5.5 in units of cm/s^2; and metallicities of -3.0 to 0.0 (in steps of 0.5 dex); cloud opacity is fixed in this model, equilibrium chemistry is assumed (alternate designations: `witte`, `witte2011`, `helling`)
- *madhusudhan*: model set from `Madhusudhan et al. (2011) <http://adsabs.harvard.edu/abs/2011ApJ...737...34M>`_ with effective temperatures of 600 K to 1700 K (steps of 50-100 K); surface gravities of 3.5 and 5.0 in units of cm/s^2; and metallicities of 0.0 to 1.0 (in steps of 0.5 dex); there are multiple cloud prescriptions for this model, equilibrium chemistry is assumed (alternate designations: `madhusudhan`)
:param: **teff**: effective temperature of the model in K (e.g. `teff` = 1000)
:param: **logg**: log10 of the surface gravity of the model in cm/s^2 units (e.g. `logg` = 5.0)
:param: **z**: log10 of metallicity of the model relative to solar metallicity (e.g. `z` = -0.5)
:param: **fsed**: sedimentation efficiency of the model (e.g. `fsed` = 'f2')
:param: **cld**: cloud shape function of the model (e.g. `cld` = 'f50')
:param: **kzz**: vertical eddy diffusion coefficient of the model (e.g. `kzz` = 2)
:param: **instrument**: instrument the model should be converted to (default = 'raw')
:param: **file**: file name for model (default = '' or generated from parameters)
:param: **folder**: folder containing file (default = '' or default folder for model set)
:param: **verbose**: give lots of feedback
:Output:
A SPLAT Spectrum object of the model with wavelength in microns and surface fluxes in F_lambda units of erg/cm^2/s/micron.
:Example:
>>> import splat
>>> mdl = splat.loadOriginalModel(model='btsettl',teff=2600,logg=4.5)
>>> mdl.info()
btsettl08 Teff=2600 logg=4.5 [M/H]=0.0 atmosphere model with the following parmeters:
Teff = 2600 K
logg = 4.5 dex
z = 0.0 dex
fsed = nc
cld = nc
kzz = eq
If you use this model, please cite <NAME>. et al. (2012, Philosophical Transactions of the Royal Society A, 370, 2765-2777)
bibcode = 2012RSPTA.370.2765A
'''
mkwargs = {}
mkwargs['ismodel'] = True
# check modelset name
mset = checkSpectralModelName(model)
if mset == False:
raise ValueError('\{} is not a valid model set name'.format(model))
mkwargs['model'] = mset
# if not a specific file, generate it
if file == '':
for k in list(SPECTRAL_MODEL_PARAMETERS.keys()):
mkwargs[k] = kwargs.get(k,SPECTRAL_MODEL_PARAMETERS[k]['default'])
if mset == 'btsettl08':
readfxn = _readBtsettl08
file = os.path.normpath(SPECTRAL_MODELS[mset]['rawfolder']+'lte{:s}-{:.1f}{:.1f}a+0.0.BT-Settl.spec.7.gz'.format(str((float(mkwargs['teff'])+0.01)/1.e5)[2:5],mkwargs['logg'],mkwargs['z']-0.0001))
elif mset == 'madhusudhan11':
readfxn = _readBurrows06
file = os.path.normpath(SPECTRAL_MODELS[mset]['rawfolder']+'{:s}_t{:.0f}_g{:.2f}_z{:.0f}_{:s}_{:s}'.format(mkwargs['cld'].upper(),int(mkwargs['teff']),float(mkwargs['logg']),10.**float(mkwargs['z']),mkwargs['kzz'].lower(),mkwargs['fsed'].lower()))
elif mset == 'saumon12':
readfxn = _readSaumon12
file = os.path.normpath(SPECTRAL_MODELS[mset]['rawfolder']+'sp_t{:.0f}g{:.0f}{:s}'.format(int(mkwargs['teff']),10.**(float(mkwargs['logg'])-2.),mkwargs['cld'].lower()))
else:
raise ValueError('\nDo not yet have {} models in loadOriginalModel'.format(mset))
# check file name
if not os.access(file, os.R_OK):
# filetmp = SPECTRAL_MODELS[mset]['rawfolder']+file
# if not os.access(filetmp, os.R_OK):
raise ValueError('Could not find model file {}'.format(file))
# else: file=filetmp
mkwargs['filename'] = os.path.basename(file)
# read in data
wave,flux = readfxn(file)
mkwargs['wave'] = wave
mkwargs['flux'] = flux
# convert to instrument - TBD
mkwargs['instrument'] = instrument
return Spectrum(**mkwargs)
def loadOriginalInterpolatedModel(model='btsettl08',teff=2000,logg=5.0,**kwargs):
'''
:Purpose:
Loads up an original model spectrum at full resolution/spectral range, interpolated by temperature and surface gravity
:Required Inputs:
None
:Optional Inputs:
same as .. `loadOriginalModel()`_
.. _`loadOriginalModel()` : api.html#splat.model.loadOriginalModel
:Output:
A SPLAT Spectrum object of the model with wavelength in microns and surface fluxes in F_lambda units of erg/cm^2/s/micron.
:Example:
>>> import splat
>>> mdl = splat.loadOriginalInterpolatedModel(model='btsettl',teff=2632,logg=4.6)
>>> mdl.info()
BT-Settl (2008) Teff=2632 logg=4.6 atmosphere model with the following parmeters:
Teff = 2632 K
logg = 4.6 dex
z = 0.0 dex
fsed = nc
cld = nc
kzz = eq
If you use this model, please cite <NAME>. et al. (2012, Philosophical Transactions of the Royal Society A, 370, 2765-2777)
bibcode = 2012RSPTA.370.2765A
'''
teffs = [100*numpy.floor(teff/100.),100*numpy.ceil(teff/100.)]
loggs = [0.5*numpy.floor(logg*2.),0.5*numpy.ceil(logg*2.)]
wt = numpy.log10(teffs[1]/teff)/numpy.log10(teffs[1]/teffs[0])
wg = (loggs[1]-logg)/(loggs[1]-loggs[0])
weights = numpy.array([wt*wg,(1.-wt)*wg,wt*(1.-wg),(1.-wt)*(1.-wg)])
weights/=numpy.sum(weights)
models = []
models.append(loadOriginalModel(model=model,teff=teffs[0],logg=loggs[0],**kwargs))
if teffs[1]==teffs[0]:
models.append(models[-1])
else:
models.append(loadOriginalModel(model=model,teff=teffs[1],logg=loggs[0],**kwargs))
if loggs[1]==loggs[0]:
models.extend(models[0:1])
else:
models.append(loadOriginalModel(model=model,teff=teffs[0],logg=loggs[1],**kwargs))
models.append(loadOriginalModel(model=model,teff=teffs[1],logg=loggs[1],**kwargs))
flx = []
for i in range(len(models[0].flux)):
val = numpy.array([numpy.log10(m.flux.value[i]) for m in models])
flx.append(10.**(numpy.sum(val*weights)))
mdl_return = models[0]
mdl_return.flux = flx*models[0].flux.unit
mdl_return.teff = teff
mdl_return.logg = logg
mdl_return.name = '{} Teff={} logg={}'.format(splat.SPECTRAL_MODELS[checkSpectralModelName(model)]['name'],teff,logg)
return mdl_return
# make model function
def makeForwardModel(parameters,data,atm=None,binary=False,duplicate=False,model=None,model1=None,model2=None,instkern=None,contfitdeg=5,return_nontelluric=False,checkplots=False,checkprefix='tmp',verbose=True,timecheck=False):
'''
parameters may contain any of the following:
- **modelparam** or **modelparam1**: dictionary of model parameters for primary if model not provided in model or model1: {modelset,teff,logg,z,fsed,kzz,cld,instrument}
- **modelparam2**: dictionary of model parameters for secondary if model not provided in model2: {modelset,teff,logg,z,fsed,kzz,cld,instrument}
- **rv** or **rv1**: radial velocity of primary
- **rv2**: radial velocity of secondary
- **vsini** or **vsini1**: rotational velocity of primary
- **vsini2**: rotational velocity of secondary
- **f21**: relative brightness of secondary to primary (f21 <= 1)
- **alpha**: exponent to scale telluric absorption
- **vinst**: instrument velocity broadening profile if instrkern not provided
- **vshift**: instrument velocity shift
- **continuum**: polynomial coefficients for continuum correction; if not provided, a smooth continuum will be fit out
- **offset**: additive flux offset, useful if there may be residual background continuum
- **offset_fraction**: additive flux offset as a fraction of the median flux, useful if there may be residual background continuum
'''
# check inputs
# timing check
timing = [time.time()]
if 'modelset' in list(parameters.keys()) and 'modelset1' not in list(parameters.keys()): parameters['modelset1'] = parameters['modelset']
if 'set' in list(parameters.keys()) and 'modelset1' not in list(parameters.keys()): parameters['modelset1'] = parameters['set']
if 'set1' in list(parameters.keys()) and 'modelset1' not in list(parameters.keys()): parameters['modelset1'] = parameters['set1']
if 'set2' in list(parameters.keys()) and 'modelset2' not in list(parameters.keys()): parameters['modelset2'] = parameters['set2']
# data
if data != None:
if isinstance(data,splat.Spectrum) == False:
raise ValueError('\nData {} must be a Spectrum object'.format(data))
# model / model parameters
if model1 == None and model != None: model1 = copy.deepcopy(model)
if model1 != None:
if isinstance(model1,splat.Spectrum) == False:
raise ValueError('\nModel for primary source {} must be a Spectrum object'.format(model1))
else:
if 'modelset1' not in list(parameters.keys()): raise ValueError('\nMust provide model parameters for primary')
if binary == True and model2 == None and 'modelset2' not in list(parameters.keys()):
parameters['modelset2'] = parameters['modelset1']
if binary == True:
if model2 != None:
if isinstance(model2,splat.Spectrum) == False:
raise ValueError('\nModel for secondary source {} must be a Spectrum object'.format(model2))
elif 'modelset2' not in list(parameters.keys()):
raise ValueError('\nMust provide model parameters for secondary')
if 'modelset1' in list(parameters.keys()):
mset1 = checkSpectralModelName(parameters['modelset1'])
if mset1 != False: parameters['modelset1'] = mset1
else: raise ValueError('Unknown model set {} for primary'.format(parameters['modelset1']))
if 'modelset2' in list(parameters.keys()):
mset2 = checkSpectralModelName(parameters['modelset2'])
if mset2 != False: parameters['modelset2'] = mset2
else: raise ValueError('Unknown model set {} for primary'.format(parameters['modelset2']))
if 'instrument' not in list(parameters.keys()): parameters['instrument'] = 'RAW'
if parameters['instrument'] not in list(splat.SPECTRAL_MODELS[parameters['modelset1']]['instruments'].keys()):
raise ValueError('Instrument {} has not been established for model {}'.format(parameters['instrument'],parameters['modelset1']))
# timing check
timing.append(time.time())
# telluric absorption
if atm != None:
if isinstance(atm,splat.Spectrum) == False:
raise ValueError('\nModel for atmosphere {} must be a Spectrum object'.format(atm))
# print(numpy.nanmin(atm.wave.value),numpy.nanmax(atm.wave.value))
# timing check
timing.append(time.time())
# establish model spectrum
if model1 != None:
mdl1 = copy.deepcopy(model1)
else:
# read in new model
mparam = {'modelset': parameters['modelset1'], 'instrument': parameters['instrument']}
for m in list(splat.SPECTRAL_MODELS[parameters['modelset1']]['default'].keys()):
if m in list(parameters.keys()): mparam[m] = parameters[m]
if '{}1'.format(m) in list(parameters.keys()): mparam[m] = parameters['{}1'.format(m)]
try:
mdl1 = getModel(**mparam)
except:
raise ValueError('\nError in creating primary model with parameters {}'.format(mparam))
# print(mparam)
# mdl1.info()
# timing check
timing.append(time.time())
# print('Reading',numpy.nanmin(mdl1.wave.value),numpy.nanmax(mdl1.wave.value))
# add in secondary if desired
if binary==True:
if duplicate == True:
mdl2 = copy.deepcopy(mdl1)
elif model2 != None:
mdl2 = copy.deepcopy(model2)
else:
mparam = {'modelset': parameters['modelset2'], 'instrument': parameters['instrument']}
for m in list(splat.SPECTRAL_MODELS[parameters['modelset2']]['default'].keys()):
if '{}2'.format(m) in list(parameters.keys()): mparam[m] = parameters['{}2'.format(m)]
if len(list(mparam.keys())) == 2:
print('Warning: no parameters provided for secondary; assuming a duplicate model')
mdl2 = copy.deepcopy(mdl1)
else:
try:
mdl2 = getModel(**mparam)
# print(mparam)
print('Model 2 original',numpy.nanmin(mdl2.wave.value),numpy.nanmax(mdl2.wave.value))
except:
raise ValueError('\nError in creating secondary model with parameters {}'.format(mparam))
# mdl2.info()
# timing check
timing.append(time.time())
# make sure everything is on the same wavelength range
if atm != None:
atm.toWaveUnit(data.wave.unit)
mdl1.toWaveUnit(data.wave.unit)
if binary == True:
mdl2.toWaveUnit(data.wave.unit)
# visualize spectra for error checking
if checkplots==True:
splot.plotSpectrum(mdl1,mdl2,colors=['k','r'],legend=['Model 1','Model 2'],file=checkprefix+'_model.pdf')
else:
if checkplots==True:
splot.plotSpectrum(mdl1,colors=['k'],legend=['Model 1'],file=checkprefix+'_model.pdf')
# timing check
timing.append(time.time())
# apply rv shift and vsini broadening the model spectrum
if 'rv' in list(parameters.keys()):
mdl1.rvShift(parameters['rv'])
elif 'rv1' in list(parameters.keys()):
mdl1.rvShift(parameters['rv1'])
if 'vshift' in list(parameters.keys()):
mdl1.rvShift(parameters['vshift'])
if 'vsini' in list(parameters.keys()):
mdl1.broaden(parameters['vsini'],method='rotation')
elif 'vsini1' in list(parameters.keys()):
mdl1.broaden(parameters['vsini1'],method='rotation')
# print('Shifted',numpy.nanmin(mdl1.wave.value),numpy.nanmax(mdl1.wave.value))
# timing check
timing.append(time.time())
if binary==True:
# print('Model 2 original',numpy.nanmin(mdl2.wave.value),numpy.nanmax(mdl2.wave.value))
if 'f2' in list(parameters.keys()):
mdl2.scale(parameters['f2'])
if 'rv2' in list(parameters.keys()):
mdl2.rvShift(parameters['rv2'])
if 'vshift' in list(parameters.keys()):
mdl2.rvShift(parameters['vshift'])
if 'vsini2' in list(parameters.keys()):
mdl2.broaden(parameters['vsini2'],method='rotation')
else:
if 'vsini' in list(parameters.keys()):
mdl2.broaden(parameters['vsini'],method='rotation')
elif 'vsini1' in list(parameters.keys()):
mdl2.broaden(parameters['vsini1'],method='rotation')
# print('Model 2 shifted',numpy.nanmin(mdl2.wave.value),numpy.nanmax(mdl2.wave.value))
# add primary and secondary back together
mdl = mdl1+mdl2
# print('Combined',numpy.nanmin(mdl.wave.value),numpy.nanmax(mdl.wave.value))
else:
mdl = mdl1
# timing check
timing.append(time.time())
# read in telluric, scale & apply
if atm != None:
# integral resample telluric profile onto mdl flux range
atmapp = copy.deepcopy(atm)
if 'vshift' in list(parameters.keys()):
atmapp.rvShift(parameters['vshift'])
if len(atmapp.flux) != len(mdl.flux):
funit = atmapp.flux.unit
# atmapp.flux = splat.integralResample(atmapp.wave.value,atmapp.flux.value,mdl.wave.value)
# print(numpy.nanmin(atmapp.wave.value),numpy.nanmax(atmapp.wave.value),numpy.nanmin(mdl.wave.value),numpy.nanmax(mdl.wave.value),)
atmapp.flux = splat.reMap(atmapp.wave.value,atmapp.flux.value,mdl.wave.value)
atmapp.flux = atmapp.flux*funit
atmapp.wave = mdl.wave
atmapp.noise = [numpy.nan for f in atmapp.flux]*funit
atmapp.variance = [numpy.nan for f in atmapp.flux]*(funit**2)
if 'alpha' in list(parameters.keys()):
atmapp.flux = [t**parameters['alpha'] for t in atmapp.flux.value]*atmapp.flux.unit
mdlt = mdl*atmapp
if checkplots==True:
splot.plotSpectrum(mdlt,mdl,colors=['r','k'],legend=['Model x Atmosphere','Model'],file=checkprefix+'_modelatm.pdf')
else: mdlt = copy.deepcopy(mdl)
# timing check
timing.append(time.time())
# correct for velocity shift from wavelength error
# if 'vshift' in list(parameters.keys()):
# mdl.rvShift(parameters['vshift'])
# mdlt.rvShift(parameters['vshift'])
# timing check
timing.append(time.time())
# resample original and telluric corrected models onto data wavelength range
funit = mdl.flux.unit
mdlsamp = copy.deepcopy(mdl)
# print(numpy.nanmin(data.wave.value),numpy.nanmax(data.wave.value))
# print(numpy.nanmin(mdl.wave.value),numpy.nanmax(mdl.wave.value))
mdlsamp.flux = splat.reMap(mdl.wave.value,mdl.flux.value,data.wave.value)
mdlsamp.flux = mdlsamp.flux*funit
mdlsamp.wave = data.wave
mdlsamp.noise = [numpy.nan for f in mdlsamp.flux]*funit
mdlsamp.variance = [numpy.nan for f in mdlsamp.flux]*(funit**2)
funit = mdlt.flux.unit
mdltsamp = copy.deepcopy(mdlt)
# mdltsamp.flux = splat.integralResample(mdlt.wave.value,mdlt.flux.value,data.wave.value)
mdltsamp.flux = splat.reMap(mdlt.wave.value,mdlt.flux.value,data.wave.value)
mdltsamp.flux = mdltsamp.flux*funit
mdltsamp.wave = data.wave
mdltsamp.noise = [numpy.nan for f in mdltsamp.flux]*funit
mdltsamp.variance = [numpy.nan for f in mdltsamp.flux]*(funit**2)
# timing check
timing.append(time.time())
if checkplots==True:
splot.plotSpectrum(mdltsamp,mdlsamp,colors=['r','k'],legend=['Model x Atmosphere','Model'],file=checkprefix+'_modelatmsamp.pdf')
# broaden by instrumental profile
if instkern != None:
mdlsamp.broaden(parameters['vinst'],kern=instkern)
mdltsamp.broaden(parameters['vinst'],kern=instkern)
elif 'vinst' in list(parameters.keys()):
mdlsamp.broaden(parameters['vinst'],method='gaussian')
mdltsamp.broaden(parameters['vinst'],method='gaussian')
if checkplots==True:
splot.plotSpectrum(mdltsamp,mdlsamp,colors=['r','k'],legend=['Model x Atmosphere','Model'],file=checkprefix+'_modelatmsampbroad.pdf')
# timing check
timing.append(time.time())
# apply flux offset (e.g. poor background subtraction)
if 'offset' in list(parameters.keys()):
funit = mdlsamp.flux.unit
mdlsamp.flux = [m+parameters['offset'] for m in mdlsamp.flux.value]*funit
funit = mdltsamp.flux.unit
mdltsamp.flux = [m+parameters['offset'] for m in mdltsamp.flux.value]*funit
if 'offset_fraction' in list(parameters.keys()):
funit = mdlsamp.flux.unit
mdlsamp.flux = [m+parameters['offset_fraction']*numpy.median(mdlsamp.flux.value) for m in mdlsamp.flux.value]*funit
funit = mdltsamp.flux.unit
mdltsamp.flux = [m+parameters['offset_fraction']*numpy.median(mdltsamp.flux.value) for m in mdltsamp.flux.value]*funit
# timing check
timing.append(time.time())
# correct for continuum
mdlcont = copy.deepcopy(mdlsamp)
mdltcont = copy.deepcopy(mdltsamp)
if 'continuum' in list(parameters.keys()):
mdlcont.flux = mdlcont.flux*numpy.polyval(parameters['continuum'],mdlcont.wave.value)
mdltcont.flux = mdlcont.flux*numpy.polyval(parameters['continuum'],mdltcont.wave.value)
else:
mdldiv = data/mdltsamp
mdldiv.smooth(pixels=20)
# NOTE: this fails if there are any nans around
pcont = numpy.polyfit(mdldiv.wave.value,mdldiv.flux.value,contfitdeg)
# f = interp1d(data.wave.value,data.flux.value)
# pcont = numpy.polyfit(mdltfinal.wave.value,f(mdltfinal.wave.value)/mdltfinal.flux.value,contfitdeg)
mdlcont.flux = mdlcont.flux*numpy.polyval(pcont,mdlcont.wave.value)
mdltcont.flux = mdltcont.flux*numpy.polyval(pcont,mdltcont.wave.value)
if checkplots==True:
mdltmp = copy.deepcopy(mdlsamp)
mdldiv = data/mdltsamp
mdltmp.scale(numpy.nanmedian(mdldiv.flux.value))
splot.plotSpectrum(mdltcont,mdlcont,data,colors=['r','k','b'],legend=['Model x Atmosphere x Continuum','Model','Data'],file=checkprefix+'_modelatmsampbroadcont.pdf')
# timing check
timing.append(time.time())
# correct for velocity shift (wavelength calibration error)
mdlfinal = copy.deepcopy(mdlcont)
mdltfinal = copy.deepcopy(mdltcont)
# if 'vshift' in list(parameters.keys()):
# mdlfinal.rvShift(parameters['vshift'])
# mdltfinal.rvShift(parameters['vshift'])
# timing check
timing.append(time.time())
# return model
mdlfinal.name = '{} model'.format(parameters['modelset1'])
mdltfinal.name = '{} model x Atmosphere'.format(parameters['modelset1'])
# print timing
if timecheck==True:
print('\n\n')
for i in range(len(timing)-1): print('Time for step {} = {} s'.format(i,timing[i+1]-timing[i]))
print('\n\n')
if return_nontelluric == True:
return mdltfinal,mdlfinal
else:
return mdltfinal
# MCMC loop
def mcmcForwardModelFit(data,param0,param_var,model=None,limits={},nwalkers=1,nsteps=100,method='standard',return_threshold=0.9,dof=0.,binary=False,duplicate=False,secondary_model=None,atm=None,report=True,report_index=10,report_each=False,file='tmp',output='all',verbose=True,**kwargs):
'''
:Purpose:
Conducts and Markov Chain Monte Carlo (MCMC) forward modeling fit of a spectrum.
This routine assumes the spectral data have already been wavelength calibrated
THIS ROUTINE IS CURRENTLY IN DEVELOPMENT
:Required Inputs:
:param data: Spectrum object containing the data to be modeled
:param param0: dictionary containing the initial parameters; allowed parameters are the same as those defined in `makeForwardModel()`_
:param param_var: dictionary containing the scales (gaussian sigmas) over which the parameters are varied at each iteration; should contain the same elements as param0. If a parameter var is set to 0, then that parameter is held fixed
:Optional Inputs:
:param: limits = {}: dictionary containing the limits of the parameters; each parameter that is limited should be matched to a two-element list defining the upper and lower bounds
:param: nwalkers = 1: number of MCMC walkers
:param: nsteps = 100: number of MCMC steps taken by each walker; the actual number of fits is nsteps x # parameters
:param: dof: degrees of freedom; if not provided, assumed to be the number of datapoints minus the number of varied parameters
:param: binary = False: set to True to do a binary model fit
:param: secondary_model = None: if binary = True, use this parameter to specify the model of the secondary
:param: model = None: Spectrum object containing the spectral model to use if assumed fixed; should be of higher resolution and wider wavelength range than the data
:param: atm = None: Spectrum object containing the atmospheric/instrumental transmission profile (e.g., `loadTelluric()`_); should be of higher resolution and wider wavelength range than the data
:param: report = True: set to True to iteratively report the progress of the fit
:param: report_index = 10: if report = True, the number of steps to provide an interim report
:param: report_each = False: set to True to save all reports separately (useful for movie making)
:param: file = 'tmp': file prefix for outputs; should include full path unless in operating in desired folder
:param: output = 'all': what to return on completion; options include:
* 'all': (default) return a list of all parameter dictionaries and chi-square values
* 'best': return only a single dictionary of the best fit parameters and the best fit chi-square value
:param: verbose = False: provide extra feedback
mcmcForwardModelFit() will also take as inputs the plotting parameters for `mcmcForwardModelReport()`_ and `plotSpectrum()`_
:Outputs:
Depending on what is set for the `output` parameter, a list or single dictionary containing model parameters, and a list or single best chi-square values.
These outputs can be fed into `mcmcForwardModelReport()`_ to visualize the best fitting model and parameters
:Example:
>>> import splat
>>> import splat.model as spmdl
>>> import astropy.units as u
>>> # read in spectrum
>>> sp = splat.Spectrum('nirspec_spectrum.txt')
>>> # read in and trim model
>>> mdl = spmdl.loadModel(model='btsettl',teff=2600,logg=5.0,raw=True)
>>> mdl.trim([numpy.min(sp.wave)-0.01*u.micron,numpy.max(sp.wave)+0.01*u.micron])
>>> # read in and trim atmospheric absorption
>>> atm = spmdl.loadTelluric(wave_range=[numpy.min(mdl.wave.value)-0.02,numpy.max(mdl.wave.value)+0.02],output='spec')
>>> # inital parameters
>>> mpar = {'rv': 0., 'vsini': 10., 'vinst': 5., 'alpha': 0.6}
>>> mvar = {'rv': 1., 'vsini': 1., 'vinst': 0.2, 'alpha': 0.05}
>>> mlim = {'vsini': [0.,500.], 'alpha': [0.,100.]}
>>> # do fit
>>> pars,chis = spmdl.mcmcForwardModelFit(sp,mdl,mpar,mvar,limits=mlim,atm=atm,nsteps=100,file='fit'')
>>> # visualize results
>>> spmdl.mcmcForwardModelReport(sp,mdl,pars,chis,file='fit',chiweights=True,plotParameters=['rv','vsini'])
'''
# check method
if method not in ['standard','best','return']: method = 'standard'
# generate first fit
if dof == 0.: dof = int(len(data.wave)-len(list(param0.keys())))
mdl = makeForwardModel(param0,data,binary=binary,duplicate=duplicate,atm=atm,model=model,model2=secondary_model)
chi0,scale = splat.compareSpectra(data,mdl)
parameters = [param0]
chis = [chi0]
acceptance = [1]
if verbose == True:
l = 'Initial guess: chi={:.0f}, dof={}'.format(chis[-1],dof)
for k in list(param_var.keys()):
if param_var[k] != 0.: l+=' , {}={:.2f}'.format(k,parameters[-1][k])
print(l)
for i in range(nsteps):
for k in list(param_var.keys()):
if param_var[k] != 0.:
param = copy.deepcopy(param0)
param[k] = numpy.random.normal(param[k],param_var[k])
# force within range with soft bounce
if k in list(limits.keys()):
if param[k] < numpy.min(limits[k]): param[k] = numpy.min(limits[k])+numpy.random.uniform()*(numpy.min(limits[k])-param[k])
if param[k] > numpy.max(limits[k]): param[k] = numpy.max(limits[k])-numpy.random.uniform()*(param[k]-numpy.max(limits[k]))
# print(numpy.nanmin(data.wave.value),numpy.nanmax(data.wave.value))
# if atm != None: print(numpy.nanmin(atm.wave.value),numpy.nanmax(atm.wave.value))
mdl = makeForwardModel(param,data,binary=binary,duplicate=duplicate,atm=atm,model=model,model2=secondary_model)
# print(numpy.nanmin(mdl.wave.value),numpy.nanmax(mdl.wave.value))
chi,scale = splat.compareSpectra(data,mdl)
# different methods
test = 2.*(stats.f.cdf(chi/chi0, dof, dof)-0.5)
if method == 'best': test = 2.*(stats.f.cdf(chi/numpy.nanmin(chis), dof, dof)-0.5)
if verbose==True: print(chi,chi0,test)
if test < numpy.random.uniform(0,1):
param0 = copy.deepcopy(param)
chi0 = chi
acceptance.append(1)
else: acceptance.append(0)
if method == 'return':
test = 2.*(stats.f.cdf(chi0/numpy.nanmin(chis), dof, dof)-0.5)
if verbose==True: print(chi0,numpy.nanmin(chis),test)
if test > return_threshold:
param0 = copy.deepcopy(parameters[numpy.argmin(chis)])
chi0 = numpy.min(chis)
if verbose == True: print('Jump made back to chi = {}'.format(chi0))
parameters.append(param0)
chis.append(chi0)
if verbose == True:
l = 'Step {}: varied {}, chi={:.0f}, dof={}'.format(i,k,chis[-1],dof)
for k in list(param_var.keys()):
if param_var[k] != 0.: l+=' , {}={:.2f}'.format(k,parameters[-1][k])
print(l)
# report where we are
if report == True and i % int(report_index) == 0 and i > 0:
# l = 'Step {}: chi={:.0f}, dof={}'.format(i,chis[-1],dof)
# for k in list(param0.keys()):
# if isinstance(parameters[-1][k],float): l+=' , {}={:.2f}'.format(k,parameters[-1][k])
# else: l+=' , {}={}'.format(k,parameters[-1][k])
# print(l)
ibest = numpy.argmin(chis)
best_parameters = parameters[ibest]
l = '\nBest chi={:.0f}, dof={}'.format(chis[ibest],dof)
for k in list(param0.keys()):
if isinstance(best_parameters[k],float): l+=' , {}={:.2f}'.format(k,best_parameters[k])
else: l+=' , {}={}'.format(k,best_parameters[k])
l = '\nCumulative acceptance = {:.2f}%, Recent acceptance = {:.2f}%'.format(100.*numpy.sum(acceptance)/len(acceptance),100.*numpy.sum(acceptance[-1*report_index:])/len(acceptance[-1*report_index:]))
print(l)
# mdl,mdlnt = makeForwardModel(parameters[-1],data,binary=binary,atm=atm,model=model,return_nontelluric=True)
# chi,scale = splat.compareSpectra(data,mdl)
# mdl.scale(scale)
# mdlnt.scale(scale)
# splot.plotSpectrum(data,mdlnt,mdl,data-mdl,colors=['k','g','r','b'],legend=['Data','Model','Model+Telluric','Difference\nChi2={:.0f}'.format(chi)],figsize=kwargs.get('figsize',[15,5]),file=file+'_interimComparison.pdf')
# mdl,mdlnt = makeForwardModel(best_parameters,data,binary=binary,atm=atm,model=model,return_nontelluric=True)
# chi,scale = splat.compareSpectra(data,mdl)
# mdl.scale(scale)
# mdlnt.scale(scale)
# splot.plotSpectrum(data,mdlnt,mdl,data-mdl,colors=['k','g','r','b'],legend=['Data','Model-Telluric','Best Model\nChi2={:.0f}'.format(chi),'Difference'],figsize=kwargs.get('figsize',[15,5]),file=file+'_bestModel.pdf')
# f = open(file+'_report.txt','w')
# f.write('steps completed = {}\n'.format(i))
# f.write('best chi^2 = {:.0f}\n'.format(chis[i]))
# f.write('degrees of freedom = {:.0f}\n'.format(dof))
# for k in list(param0.keys()): f.write('{} = {:.2f}\n'.format(k,best_parameters[k]))
# f.close()
final_parameters = {}
for k in list(param0.keys()):
vals = []
for i in range(len(parameters)): vals.append(parameters[i][k])
final_parameters[k] = vals
mcmcForwardModelReport(data,final_parameters,chis,dof=dof,atm=atm,file=file,binary=binary,duplicate=duplicate,verbose=verbose,**kwargs)
# identify best model
ibest = numpy.argmin(chis)
best_chi = chis[ibest]
best_parameters = parameters[ibest]
# reformat parameters
final_parameters = {}
for k in list(param0.keys()):
vals = []
for i in range(len(parameters)): vals.append(parameters[i][k])
final_parameters[k] = vals
if report == True:
l = 'Best chi={:.0f}, dof={}'.format(best_chi,dof)
for k in list(best_parameters.keys()):
if isinstance(best_parameters[k],float): l+=' , {}={:.2f}'.format(k,best_parameters[k])
else: l+=' , {}={}'.format(k,best_parameters[k])
print(l)
mcmcForwardModelReport(data,final_parameters,chis,dof=dof,atm=atm,file=file,binary=binary,duplicate=duplicate,verbose=verbose,**kwargs)
# mdl,mdlnt = makeForwardModel(best_parameters,data,binary=binary,atm=atm,model=model,return_nontelluric=True)
# chi0,scale = splat.compareSpectra(data,mdl)
# mdl.scale(scale)
# mdlnt.scale(scale)
# splot.plotSpectrum(data,mdlnt,mdl,data-mdl,colors=['k','g','r','b'],legend=['Data','Model-Telluric','Best Model\nChi2={:.0f}'.format(best_chi),'Difference'],figsize=figsize,file=file+'_bestModel.pdf')
# burn off beginning of chain
# burned_parameters = parameters[int(burn*len(parameters)):]
# burned_chis = chis[int(burn*len(chis)):]
# if 'burn' in output.lower():
# parameters = burned_parameters
# chis = burned_chis
# correct for barycentric motion
# if isinstance(vbary,u.quantity.Quantity):
# vb = vbary.to(u.km/u.s).value
# else:
# vb = copy.deepcopy(vbary)
# if 'rv' in list(param0.keys()):
# final_parameters['rv'] = [r+vb for r in final_parameters['rv']]
# best_parameters['rv']+=vb
# if 'rv1' in list(param0.keys()):
# final_parameters['rv1'] = [r+vb for r in final_parameters['rv1']]
# best_parameters['rv1']+=vb
# if 'rv2' in list(param0.keys()):
# final_parameters['rv2'] = [r+vb for r in final_parameters['rv2']]
# best_parameters['rv2']+=vb
# return values
if 'best' in output.lower():
return best_parameters, best_chi
else:
return final_parameters, chis
def mcmcForwardModelReport(data,parameters,chis,burn=0.25,dof=0,plotChains=True,plotBest=True,plotMean=True,plotCorner=True,plotParameters=None,writeReport=True,vbary=0.,file='tmp',atm=None,model=None,model2=None,chiweights=False,binary=False,duplicate=False,verbose=True):
'''
:Purpose:
Plotting and fit analysis routine for `mcmcForwardModelFit()`_
:Required Inputs:
:param data: Spectrum object containing the data modeled
:param parameters: dictionary containing the parameters from the fit; each parameter should be linked to a array
:param chis: list of the chi-square values (or equivalent statistic) that match the parameter arrays
:Optional Inputs:
:param: atm = None: Spectrum object containing the atmospheric/instrumental transmission profile (e.g., `loadTelluric()`_)
:param: burn = 0.25: initial fraction of parameters to throw out ("burn-in")
:param: dof = 0: degrees of freedom; if not provided, assumed to be the number of datapoints minus the number of varied parameters
:param: binary = False: set to True if a binary model fit was done
:param: duplicate = False: set to True if the secondary spectrum has same model parameters as primary
:param: vbary = 0.: set to a velocity (assumed barycentric) to add to rv values
:param: model = None: Spectrum object containing the primary spectral model; should be of higher resolution and wider wavelength range than the data
:param: model2 = None: Spectrum object containing the secondary spectral model; should be of higher resolution and wider wavelength range than the data
:param: plotParameters = None: array of the parameters to plot, which should be keys in teh parameters input dictionary; if None, all of the parameters are plot
:param: plotChains = True: set to True to plot the parameter & chi-square value chains
:param: plotBest = True: set to True to plot the best fit model
:param: plotMean = True: set to True to plot the mean parameter model
:param: plotCorner = True: set to True to plot a corner plot of parameters (requires corner.py package)
:param: writeReport = True: set to True to write out best and average parameters to a file
:param: chiweights = False: apply chi-square weighting for determining mean parameter values
:param: file = 'tmp': file prefix for outputs; should include full path unless in operating in desired folder
:param: verbose = False: provide extra feedback
mcmcForwardModelReport() will also take as inputs the plotting parameters for `plotSpectrum()`_
:Outputs:
Depending on the flags set, various plots showing the derived parameters and best fit model for `mcmcForwardModelFit()`_
:Example:
>>> import splat
>>> import splat.model as spmdl
>>> import astropy.units as u
>>> # read in spectrum
>>> sp = splat.Spectrum('nirspec_spectrum.txt')
>>> # read in and trim model
>>> mdl = spmdl.loadModel(model='btsettl',teff=2600,logg=5.0,raw=True)
>>> mdl.trim([numpy.min(sp.wave)-0.01*u.micron,numpy.max(sp.wave)+0.01*u.micron])
>>> # read in and trim atmospheric absorption
>>> atm = spmdl.loadTelluric(wave_range=[numpy.min(mdl.wave.value)-0.02,numpy.max(mdl.wave.value)+0.02],output='spec')
>>> # inital parameters
>>> mpar = {'rv': 0., 'vsini': 10., 'vinst': 5., 'alpha': 0.6}
>>> mvar = {'rv': 1., 'vsini': 1., 'vinst': 0.2, 'alpha': 0.05}
>>> mlim = {'vsini': [0.,500.], 'alpha': [0.,100.]}
>>> # do fit
>>> pars,chis = spmdl.mcmcForwardModelFit(sp,mdl,mpar,mvar,limits=mlim,atm=atm,nsteps=100,file='fit'')
>>> # visualize results
>>> spmdl.mcmcForwardModelReport(sp,mdl,pars,chis,file='fit',chiweights=True,plotParameters=['rv','vsini'])
'''
par = copy.deepcopy(parameters)
chi = copy.deepcopy(chis)
nval = len(chis)
ns = copy.deepcopy(data)
ns.flux = ns.noise
ns2 = copy.deepcopy(data)
ns2.flux = -1.*ns.noise
# burn first X% of chains
if burn != 0. and burn < 1.:
for k in list(par.keys()): par[k] = par[k][int(nval*burn):]
chi = chi[int(nval*burn):]
nval = len(chi)
# apply weighting function
weights = numpy.ones(nval)
if chiweights==True:
if dof == 0: dof = int(len(data.wave)-len(list(par.keys())))
weights = [stats.f.sf(c/numpy.nanmin(chi),dof,dof) for c in chi]
weights = numpy.array(weights)
weights=weights/numpy.nansum(weights)
# correct velocities for barycentric motion
if isUnit(vbary):
vb = vbary.to(u.km/u.s).value
else:
vb = copy.deepcopy(vbary)
if 'rv' in list(par.keys()): par['rv'] = [r+vb for r in par['rv']]
if 'rv1' in list(par.keys()): par['rv1'] = [r+vb for r in par['rv1']]
if 'rv2' in list(par.keys()):
par['rv2'] = [r+vb for r in par['rv2']]
par['rv1-rv2'] = numpy.array(par['rv1'])-numpy.array(par['rv2'])
par['rv2-rv1'] = numpy.array(par['rv2'])-numpy.array(par['rv1'])
# best parameters
i = numpy.argmin(chi)
best_parameters = {}
for k in list(par.keys()): best_parameters[k] = par[k][i]
if verbose == True:
print('\nBest Parameter Values:')
for k in list(par.keys()): print('\t{} = {}'.format(k,best_parameters[k]))
print('\tMinimum chi^2 = {}'.format(numpy.nanmin(chi)))
# plot best model
if plotBest==True:
plt.clf()
mdl,mdlnt = makeForwardModel(best_parameters,data,binary=binary,atm=atm,model=model,model2=model2,duplicate=duplicate,return_nontelluric=True)
chi0,scale = splat.compareSpectra(data,mdl)
mdl.scale(scale)
mdlnt.scale(scale)
if atm == None:
splot.plotSpectrum(data,mdl,data-mdl,ns,ns2,colors=['k','r','b','grey','grey'],linestyles=['-','-','-','--','--'],legend=['Data','Model',r'Difference $\chi^2$='+'{:.0f}'.format(chi0),'Noise'],figsize=[15,5],yrange=[-2.*numpy.nanmedian(ns.flux.value),1.5*numpy.nanmax(mdl.flux.value)],file=file+'_bestModel.pdf')
else:
splot.plotSpectrum(data,mdl,mdlnt,data-mdl,ns,ns2,colors=['k','r','r','b','grey','grey'],linestyles=['-','-','--','-','--','--'],legend=['Data','Model x Telluric','Model',r'Difference $\chi^2$='+'{:.0f}'.format(chi0),'Noise'],figsize=[15,5],yrange=[-3.*numpy.nanmedian(ns.flux.value),1.5*numpy.nanmax(mdl.flux.value)],file=file+'_bestModel.pdf')
# mean parameters
mean_parameters = {}
mean_parameters_unc = {}
for k in list(par.keys()):
try:
mean_parameters[k] = numpy.nansum(numpy.array(par[k])*weights)
mean_parameters_unc[k] = numpy.sqrt(numpy.nansum((numpy.array(par[k])**2)*weights)-mean_parameters[k]**2)
except:
pass
if verbose == True:
print('\nMean Parameter Values:')
for k in list(mean_parameters.keys()): print('\t{} = {}+/-{}'.format(k,mean_parameters[k],mean_parameters_unc[k]))
# plot mean model
if plotMean==True:
plt.clf()
mdl,mdlnt = makeForwardModel(mean_parameters,data,binary=binary,atm=atm,model=model,model2=model2,duplicate=duplicate,return_nontelluric=True)
chi0,scale = splat.compareSpectra(data,mdl)
mdl.scale(scale)
mdlnt.scale(scale)
if atm == None:
splot.plotSpectrum(data,mdl,data-mdl,ns,ns2,colors=['k','r','b','grey','grey'],linestyles=['-','-','-','--','--'],legend=['Data','Model',r'Difference $\chi^2$='+'{:.0f}'.format(chi0),'Noise'],figsize=[15,5],yrange=[-2.*numpy.nanmedian(ns.flux.value),1.5*numpy.nanmax(mdl.flux.value)],file=file+'_meanModel.pdf')
else:
splot.plotSpectrum(data,mdl,mdlnt,data-mdl,ns,ns2,colors=['k','r','r','b','grey','grey'],linestyles=['-','-','--','-','--','--'],legend=['Data','Model x Telluric','Model',r'Difference $\chi^2$='+'{:.0f}'.format(chi0),'Noise'],figsize=[15,5],yrange=[-3.*numpy.nanmedian(ns.flux.value),1.5*numpy.nanmax(mdl.flux.value)],file=file+'_meanModel.pdf')
# print(plotParameters,toplot.keys(),best_parameters.keys(),mean_parameters.keys())
# summarize results to a text file
if writeReport==True:
f = open(file+'_report.txt','w')
f.write('Last Parameter Values (step {}):'.format(len(chi)))
for k in list(par.keys()): f.write('\n\t{} = {}'.format(k,par[k][-1]))
f.write('\n\tchi^2 = {}'.format(chi[-1]))
f.write('\n\nBest Parameter Values:')
for k in list(best_parameters.keys()): f.write('\n\t{} = {}'.format(k,best_parameters[k]))
f.write('\n\tchi^2 = {}'.format(numpy.nanmin(chi)))
f.write('\n\nMean Parameter Values:')
for k in list(mean_parameters.keys()): f.write('\n\t{} = {}+/-{}'.format(k,mean_parameters[k],mean_parameters_unc[k]))
f.close()
# prep plotting
if plotParameters == None:
plotParameters = list(mean_parameters.keys())
toplot = {}
for k in plotParameters:
if k in list(par.keys()):
if numpy.nanstd(par[k]) > 0.:
if k in list(mean_parameters.keys()):
if numpy.isfinite(mean_parameters_unc[k]):
toplot[k] = par[k]
else:
print('\nWarning: parameter {} not in MCMC parameter list; ignoring'.format(k))
# plot chains
if plotChains==True:
plt.clf()
plt.figure(figsize=(15,3*(len(list(toplot.keys()))+1)))
for i,k in enumerate(list(toplot.keys())):
# f,(ax1,ax2) = plt.subplots(1,2,sharey=True)
# ax1.plot(range(len(toplot[k])),toplot[k],'k-')
# ax1.plot([0,len(toplot[k])],[best_parameters[k],best_parameters[k]],'b-')
# ax1.plot([0,len(toplot[k])],[mean_parameters[k],mean_parameters[k]],'g--')
# ax1.set_ylabel(str(k))
# ax1.set_xlim([0,len(toplot[k])])
# ax1.legend(['Best Value = {:.3f}'.format(best_parameters[k]),'Mean Value = {}'.format(mean_parameters[k])])
# ax2.hist(toplot[k], bins=numpy.min([20,int(numpy.sqrt(len(toplot[k])))]), orientation="horizontal")
# f.subplots_adjust(hspace=0)
# plt.setp([a.get_xticklabels() for a in f.axes[:-1]], visible=False)
cplt = plt.subplot(len(list(toplot.keys()))+1,2,2*i+1)
plt.plot([0,len(toplot[k])],[best_parameters[k],best_parameters[k]],'b-')
plt.plot([0,len(toplot[k])],[mean_parameters[k],mean_parameters[k]],'g--')
plt.plot(range(len(toplot[k])),toplot[k],'k-')
plt.ylabel(str(k))
plt.xlim([0,len(toplot[k])])
plt.legend(['Best Value = {:.3f}'.format(best_parameters[k]),'Mean Value = {:.3f}'.format(mean_parameters[k])+r'$\pm$'+'{:.3f}'.format(mean_parameters_unc[k])])
hplt = plt.subplot(len(list(toplot.keys()))+1,2,2*i+2)
n,bins,patches = hplt.hist(toplot[k], bins=20, orientation="horizontal")
plt.plot([0,numpy.max(n)],[mean_parameters[k],mean_parameters[k]],'k-')
plt.plot([0,numpy.max(n)],[mean_parameters[k]-mean_parameters_unc[k],mean_parameters[k]-mean_parameters_unc[k]],'k--')
plt.plot([0,numpy.max(n)],[mean_parameters[k]+mean_parameters_unc[k],mean_parameters[k]+mean_parameters_unc[k]],'k--')
plt.subplot(len(list(toplot.keys()))+1,1,i+2)
plt.plot([0,len(toplot[k])],[numpy.nanmin(chi),numpy.nanmin(chi)],'b--')
plt.plot(range(len(toplot[k])),chi,'k-')
plt.ylabel(r'$\chi^2$')
plt.xlim([0,len(toplot[k])])
plt.legend(['Minimum chi2 = {:.1f}'.format(numpy.nanmin(chi))])
plt.savefig(file+'_chains.pdf')
# plot corner
if plotCorner==True:
try:
import corner
except:
print('\n\n*** You must install corner to make corner plots: https://github.com/dfm/corner.py ***\n\n')
else:
plt.clf()
pd = | pandas.DataFrame(toplot) | pandas.DataFrame |
import xgboost as xgb
import graphviz
import numpy as np
import pandas as pd
import random
import matplotlib
import textwrap
import scipy.spatial.distance as ssd
from scipy.stats import ks_2samp
from scipy.stats import entropy
import warnings
from sklearn import tree
from sklearn.manifold import TSNE
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import export_graphviz
from sklearn.cluster import KMeans
from scipy.cluster.hierarchy import dendrogram, linkage
from sklearn.preprocessing import LabelEncoder, OneHotEncoder, label_binarize
from sklearn.tree import export_graphviz
from sklearn.metrics import precision_score, precision_recall_curve, average_precision_score
from matplotlib import pyplot as plt
import re
import math
from os import listdir
from bokeh.layouts import gridplot
from bokeh.models import LinearAxis,FactorRange,Range1d,LabelSet,Label,ColumnDataSource,HoverTool,WheelZoomTool,PanTool,BoxZoomTool,ResetTool,SaveTool,BasicTicker,ColorBar,LinearColorMapper,PrintfTickFormatter,DataSource
from bokeh.palettes import brewer,inferno,magma,viridis,grey,Category10
from bokeh.plotting import figure, show, output_file
from bokeh.transform import transform,factor_cmap
from bokeh.io import export_png
from graphviz import Source
from itertools import cycle
from sklearn.decomposition import PCA
import rpy2.robjects.packages as rpackages
import rpy2.robjects as robjects
import statsmodels.api as sm
import re
from PIL import Image
Image.warnings.simplefilter('ignore', Image.DecompressionBombWarning)
warnings.filterwarnings('ignore', category=PendingDeprecationWarning)
warnings.filterwarnings('ignore', category=ResourceWarning)
warnings.filterwarnings('ignore', category=FutureWarning)
rstats = rpackages.importr('stats')
warnings.simplefilter("error")
#####print table#####
def ppt(table,lines=30,maxWidth = 18,minWidth=10,keepDecimal=2,lineWidth=170):
if table.shape[0]<1000:
lines=table.shape[0]
if(minWidth<5):
minWidth=5
if(maxWidth<minWidth):
print("minWidth cannot large than maxWidth")
if(lines == -1):
lines = table.shape[0]
dicType = generateTypeDic(table)
dictWidth = generateWidthDic(table,lines,maxWidth,keepDecimal,minWidth)
strTemp='|'
for col in table:
if(dictWidth[col]<minWidth):
strTemp = strTemp + truncateString(str(col).upper(),minWidth)
else:
strTemp = strTemp + truncateString(str(col).upper(),dictWidth[col])
string = textwrap.fill(text=strTemp,width=lineWidth)
print(string)
count = 0
for index in range(0,table.shape[0]):
if(count<lines):
strTemp='|'
for col in table:
if(dicType[col] is 'str'):
strTemp = strTemp + truncateString(str(table[col].iloc[index]),dictWidth[col])
else:
strTemp = strTemp + truncateNumber(decimalIdentical(table[col].iloc[index],keepDecimal),dictWidth[col])
string = textwrap.fill(text=strTemp,width=lineWidth)
print(string)
else:
break
count +=1
def generateTypeDic(table):
dicType={}
for col in table:
if(type(table[col].iloc[0]) is np.float64):#if type is np.float64 and the element is NaN, the type is defined as np.float64
dicType[col]='float'
elif(type(table[col].iloc[0]) is np.int32):
dicType[col]='int'
else:
dicType[col]='str' #if type is str and the element is NaN, the type is defined as float, so that it will be asigned value as str.
return dicType
def generateWidthDic(table,lines,maxWidth,keepDecimal=2,minWidth=5):
dicWidth={}
for col in table:
width = minWidth
colType = generateTypeDic(table)[col]
if(colType is 'str'):
for index in range(0,lines):
lenth = len(str(table[col].iloc[index]))
if(lenth > width):
width = lenth
if(width > maxWidth):
width = maxWidth
elif(colType is 'float' or colType is 'int'):
for index in range(0,lines):
lenth = len(decimalIdentical(table[col].iloc[index],keepDecimal))
if(lenth > width):
width = lenth
if(width > max(9,minWidth)):
width = max(9,minWidth)
dicWidth[col] = width
return dicWidth
def decimalIdentical(flt,kd):
if(np.isnan(flt)):
return str(flt)
flStr = str(round(flt,kd))
try:
dotIndex = len(flStr)-flStr.index('.')-1
except ValueError as ve:
flStr = flStr+'.'
dotIndex = 0
for i in range(dotIndex,kd):
flStr = flStr + '0'
return flStr
def truncateNumber(string, length):
strTemp=''
if(len(string)<9):
strTemp=string
for _ in range(len(string),length+2):
strTemp=" "+strTemp
strTemp=strTemp
else:
scientificNotation='%.2E' % Decimal(string)
strTemp=str(scientificNotation)
for _ in range(len(strTemp),length+2):
strTemp=" "+strTemp
if(len(strTemp)!=length+2):
print('truncateNumber is wrong!'+str(strTemp)+' '+str(length))
else:
return strTemp+'| '
def truncateString(string, length):
strTemp=''
if(len(string)<=length):
strTemp=string
for _ in range(len(string),length+2):
strTemp=" "+strTemp
strTemp=strTemp
else:
strTemp=string[0:length]
strTemp=strTemp+".."
if(len(strTemp)!=length+2):
print('truncateString is wrong!'+str(strTemp))
else:
return strTemp+'| '
def generate_reordered_features(megadata_validation,numeric_features_validation,basic_info_features,simple_scale):
clustering_data_validation = megadata_validation.copy()
clustering_data_validation = scale_matrix(clustering_data_validation[numeric_features_validation],isrow=False,simple_scale=simple_scale)
corr_validation_DF = pd.DataFrame(clustering_data_validation,columns=numeric_features_validation).corr('spearman')
distance_validation_Matrix = corr_validation_DF.values
for i in range(0,len(distance_validation_Matrix)):
distance_validation_Matrix[i]=1-distance_validation_Matrix[i]
distance_validation_Matrix = ssd.squareform(distance_validation_Matrix)
linked = linkage(distance_validation_Matrix,'ward','euclidean',True)
labelList = corr_validation_DF.index
featureDict= {i:[corr_validation_DF.index[i]] for i in range(0,len(corr_validation_DF.index))}
for i in range(0,len(linked)):
index = i+linked.shape[0]+1
firstList = featureDict[linked[i][0]]
for j in featureDict[linked[i][1]]:
firstList.append(j)
if(len(firstList)!=linked[i][3]):
print("the length is not equal")
featureDict[index]=firstList
featureList=featureDict[linked.shape[0]*2]
for i in range(len(basic_info_features)):
featureList.append(basic_info_features[i])
return featureList
def prepare_scaled_data(layer1_df,layer2_df,layer3_df,reordered_feature_list,simple_scale=False):
layer1_mx = layer1_df.copy()
layer1_mx = layer1_mx.replace(0,1E-7)
layer1_mx = layer1_mx.fillna(1E-8)[reordered_feature_list]
layer2_mx = layer2_df.copy()
layer2_mx = layer2_mx.replace(0,1E-7)
layer2_mx = layer2_mx.fillna(1E-8)[reordered_feature_list]
layer3_mx = layer3_df.copy()
layer3_mx = layer3_mx.replace(0,1E-7)
layer3_mx = layer3_mx.fillna(1E-8)[reordered_feature_list]
layer3_mx = layer3_mx.to_numpy()
layer2_mx = layer2_mx.to_numpy()
layer1_mx = layer1_mx.to_numpy()
layer1_mx = scale_matrix(layer1_mx,isrow=False,simple_scale=simple_scale)
layer2_mx = scale_matrix(layer2_mx,isrow=False,simple_scale=simple_scale)
layer3_mx = scale_matrix(layer3_mx,isrow=False,simple_scale=simple_scale)
return layer1_mx,layer2_mx,layer3_mx
def insert_values_between_original_data(for_image_data_matrix):
new_matrix = []
new_matrix.append(for_image_data_matrix[0])
for i in range(1,len(for_image_data_matrix)):
new_matrix.append((for_image_data_matrix[i-1]+for_image_data_matrix[i])/2)
new_matrix.append(for_image_data_matrix[i])
return np.array(new_matrix)
def plot_colorful_CNN_images(layer1_mx,layer2_mx,layer3_mx,cate,path,interpolation_row=0,interpolation_col=0):
for i in list(set(cate)):
ll=[j for j in range(len(cate)) if cate[j] == i]
for_image_data_matrix1 = layer1_mx[ll]
for_image_data_matrix2 = layer2_mx[ll]
for_image_data_matrix3 = layer3_mx[ll]
category = i
for i in range(interpolation_row):
for_image_data_matrix1 = insert_values_between_original_data(for_image_data_matrix1)
for_image_data_matrix2 = insert_values_between_original_data(for_image_data_matrix2)
for_image_data_matrix3 = insert_values_between_original_data(for_image_data_matrix3)
for i in range(interpolation_col):
for_image_data_matrix1 = insert_values_between_original_data(for_image_data_matrix1.T).T
for_image_data_matrix2 = insert_values_between_original_data(for_image_data_matrix2.T).T
for_image_data_matrix3 = insert_values_between_original_data(for_image_data_matrix3.T).T
colorful = [[[for_image_data_matrix1[j][l],for_image_data_matrix2[j][l],for_image_data_matrix3[j][l]] for l in range(0,len(for_image_data_matrix1[j]))] for j in range(0,len(for_image_data_matrix1))]
matplotlib.image.imsave(path+'/combined_'+category+'.png',colorful)
matplotlib.image.imsave(path+'/layer3_'+category+'.png',for_image_data_matrix3)
matplotlib.image.imsave(path+'/layer2_'+category+'.png',for_image_data_matrix2)
matplotlib.image.imsave(path+'/layer1_'+category+'.png',for_image_data_matrix1)
def plot_colorful_images_wrapper(megadata_temp1,megadata_temp2,megadata_temp3,numeric_cols,image_col,interpolation_row,interpolation_col,path,generate_reordered_indices,simple_scale=True):
reordered_features = generate_reordered_features(megadata_temp1,numeric_cols,[],simple_scale)
reordered_indices = generate_reordered_indices(megadata_temp1,reordered_features)
reordered_df1 = pd.DataFrame(megadata_temp1, index=reordered_indices)[reordered_features+[image_col]]
reordered_df2 = pd.DataFrame(megadata_temp2, index=reordered_indices)[reordered_features+[image_col]]
reordered_df3 = pd.DataFrame(megadata_temp3, index=reordered_indices)[reordered_features+[image_col]]
mx1,mx2,mx3 = prepare_scaled_data(reordered_df1,reordered_df2,reordered_df3,reordered_features,simple_scale)
plot_colorful_CNN_images(mx1,mx2,mx3,reordered_df1[image_col].values.tolist(),path,interpolation_row,interpolation_col)
#####Select certain rows from dataFrame based on the combined conditions related to index1 and index2#####
def combined_conditions_filter(condition_map,data_frame,index1,index2):
dataFrame=data_frame.copy()
dataFrame[index1] = dataFrame[index1].astype(str)
dataFrame[index2] = dataFrame[index2].astype(str)
dataFrame['filter'] = dataFrame[index1] + '***' + dataFrame[index2]
lst = list(str(key)+'***'+str(value) for key,value in condition_map.items())
subComLevData = dataFrame[dataFrame['filter'].isin(lst)]
del subComLevData['filter']
return subComLevData
#Unit vectors transformation of a Matrix.
def generate_unit_modules(mx, isrow=True, is_scale=True, simple_scale=True):
test = mx.copy()
if(mx.min()<0):
is_scale=True
print('there is a negative value, the matrix is auto scaled')
if(is_scale):
test=scale_matrix(test,isrow=isrow,simple_scale=simple_scale)
if(isrow):
for i in range(0,len(test)):
if(test[i].sum()!=0):
test[i] = test[i]/test[i].sum()
else:
test_t = np.transpose(test)
for i in range(0,len(test_t)):
if(test_t[i].sum()!=0):
test_t[i] = test_t[i]/test_t[i].sum()
test = np.transpose(test_t)
return test
#return the top or bottom n indices of a list
def topOrBottomN(lst,n,isabs=False,isbottom=False):
if (isabs):
sortList = []
for i in range(0,len(lst)):
sortList.append(abs(lst[i]))
else:
sortList = lst
sortDF = pd.DataFrame({'sort':sortList})
sortDF['index'] = sortDF.index
sortDF = sortDF.sort_values(by='sort', ascending=isbottom)
indexList = sortDF['index'].tolist()
return indexList[0:n]
#scale matrix based on row or col by simple method or median_transform
def scale_matrix(tst,isrow=True,simple_scale=True):
test=np.copy(tst)
if(simple_scale):
if(isrow):
for i in range(0,len(test)):
if(test[i].max()==test[i].min()):
if(test[i].max()==0):
test[i]=np.zeros(len(test[i]))
else:
test[i]=np.ones(len(test[i]))
else:
test[i] = (test[i]-test[i].min())/(test[i].max() - test[i].min())
else:
test_t = np.transpose(test)
for i in range(0,len(test_t)):
if(test[i].max()==test[i].min()):
if(test[i].max()==0):
test[i]=np.zeros(len(test[i]))
else:
test[i]=np.ones(len(test[i]))
else:
test_t[i] = (test_t[i]-test_t[i].min())/(test_t[i].max() - test_t[i].min())
test = np.transpose(test_t)
else:
if(isrow):
for i in range(0,len(test)):
test[i] = median_transform(test[i],1,0)
else:
test_t = np.transpose(test)
for i in range(0,len(test_t)):
test_t[i] = median_transform(test[:,i],1,0)
test = np.transpose(test_t)
return test
#sort data frame
def rrd(DF,sort_column='ID',asc=True,reidx=True):
new_DF=DF.copy()
new_DF=DF.sort_values(by=sort_column, ascending=True)
if(reidx):
new_DF=new_DF.reset_index(drop=True)
return new_DF
# dataframe to matrix
def dtm(data_frame,matrix_features,sort_column='ID',asc=True):
data_frame_copy = data_frame.copy()
data_frame_copy = data_frame_copy.sort_values(by=sort_column, ascending=asc)
mtx = data_frame_copy[matrix_features].values
return mtx
# matrix to dataframe
def mtd(mtx,numeric_features, data_frame=pd.DataFrame(),basic_info_feautes=[], sort_column='ID',asc=True):
DF = pd.DataFrame(mtx,columns=numeric_features)
if((data_frame.size>0)&(len(basic_info_feautes)>0)):
DF[basic_info_feautes] = rrd(data_frame,sort_column,asc).reset_index(drop=True)[basic_info_feautes]
return rrd(DF,sort_column)
def scale_transform1(lst1,lst2,scale,lowerbound):
if(len(lst1)!=len(lst2)):
print('lst1 and lst2 are not in the same size')
return None
if(lst2.min()==lst2.max()):
if(lst2.min()==0):
return [0 for _ in range(len(lst))]
else:
return [1 for _ in range(len(lst))]
return ((lst1-lst2.min())/(lst2.max()-lst2.min()))*scale+lowerbound
def scale_transform2(lst1,lst2,scale,lowerbound):
if(len(lst1)!=len(lst2)):
print('lst1 and lst2 are not in the same size')
return None
if(min(lst1.min(),lst2.min())<0):
print('warning: there is a negative value')
return None
if(lst2.max()==0):
print('error: the max value is equal to zero')
return None
return (lst1/lst2.max())*scale+lowerbound
#scale list
def median_transform(lst,scale,lowerbound):
if(len(set(lst))<2):
return np.full(len(lst), (scale+lowerbound)/2)
if(lst.min()==lst.max()):
if(lst.min()==0):
return [0 for _ in range(len(lst))]
else:
return [1 for _ in range(len(lst))]
else:
scaled_list=scale_transform1(lst,lst,scale,lowerbound)
scaled_list = scaled_list/np.median(scaled_list)
lower_list=np.array([i for i in scaled_list if i<=1]).copy()
upper_list=np.array([i for i in scaled_list if i>1]).copy()
for i in range(len(scaled_list)):
if(scaled_list[i]<=1):
if(np.ptp(lower_list)==0):
scaled_list[i]=0
else:
scaled_list[i]=scale_transform1(scaled_list[i],lower_list,0.5*(scale+lowerbound),lowerbound)
else:
scaled_list[i]=scale_transform2(scaled_list[i],upper_list,0.5*(scale+lowerbound),0.5*(scale+lowerbound))
return scaled_list
def unify_df(rna_df,numeric_features,id_col,other_categorical_features):
rna_scaled_df=rna_df.copy()
rna_scaled_df['temp_indices_within_this_function']=rna_scaled_df[id_col]
temp_mx = dtm(rna_scaled_df,numeric_features,sort_column='temp_indices_within_this_function')
temp_mx = generate_unit_modules(temp_mx, isrow=True, is_scale=True, simple_scale=True)
rna_scaled_df = mtd(temp_mx,numeric_features,rna_scaled_df,other_categorical_features+['temp_indices_within_this_function'], sort_column='temp_indices_within_this_function',asc=True)
rna_scaled_df.index=rna_scaled_df['temp_indices_within_this_function']
rna_scaled_df[id_col]=rna_scaled_df['temp_indices_within_this_function']
del rna_scaled_df['temp_indices_within_this_function']
temp_df=rna_df.copy()
temp_df.index=temp_df[id_col]
#control the scale of the mean by dividing by len(numeric_features)
temp_df = pd.DataFrame(scale_transform1(temp_df[numeric_features].T.describe().T['mean'],temp_df[numeric_features].T.describe().T['mean'],1,0)/len(numeric_features))
temp_df.columns=['mean_from_unify_df']
rna_scaled_df=pd.merge(rna_scaled_df,temp_df,left_index=True,right_index=True)
rna_scaled_df=rna_scaled_df.reset_index(drop=True)
new_numeric_features=numeric_features+['mean_from_unify_df']
return rna_scaled_df,new_numeric_features
#####find distict items in two lists#####
def findDistinct(ind1,ind2):
return (list(np.setdiff1d(ind1, ind2)),list(np.setdiff1d(ind2, ind1)))
def handle_unbalanced_dataset(df,numeric_features,label,id_column):
max_count = df.groupby([label], as_index=False).count()[[label,id_column]].sort_values(by=id_column, ascending=False).iloc(0)[0][1]
iter_list = df.groupby([label], as_index=False).count()[[label,id_column]].sort_values(by=id_column, ascending=False).values
add_sample_size_dict={i[0]:max_count-i[1] for i in iter_list}
new_DF=df.copy()
num=0
for k,v in add_sample_size_dict.items():
sample_size = df[df[label]==k].shape[0]
sample_matrix = df[df[label]==k][numeric_features].values
new_matrix=[]
for i in range(v):
two_samples_list = random.sample(range(sample_size),2)
new_sample=(sample_matrix[two_samples_list[0]]+sample_matrix[two_samples_list[1]])/2
new_matrix.append(new_sample)
new_matrix = np.array(new_matrix)
if(len(new_matrix)==0):
continue
temp_DF=pd.DataFrame(new_matrix,columns=numeric_features)
temp_DF[id_column]=np.array(['fakeid'+str(j) for j in range(num,num+temp_DF.shape[0])])
temp_DF[label]=k
num=num+temp_DF.shape[0]
new_DF = new_DF.append(temp_DF, sort=False)
new_DF.index = new_DF[id_column]
return new_DF
def cross_validation_split_with_unbalance_data(df,numeric_features,label='Category',id_column='PlateID',test_size=0.2,handle_unbalance=True):
iter_list = df.groupby([label], as_index=False).count()[[label,id_column]].sort_values(by=id_column, ascending=False).values
select_size_dict={i[0]:int(test_size*i[1]) for i in iter_list}
sample_size_dict={i[0]:i[1] for i in iter_list}
columns_list=df.columns
train_matrix=[]
test_matrix=[]
train_index=[]
test_index=[]
for k,v in select_size_dict.items():
sample_matrix = df[df[label]==k].values
selected_list = random.sample(range(sample_size_dict[k]),v)
unselected_list = findDistinct(selected_list,list(range(sample_size_dict[k])))[1]
for idx in selected_list:
test_matrix.append(sample_matrix[idx])
test_index.append(df[df[label]==k].iloc[idx][id_column])
for idx in unselected_list:
train_matrix.append(sample_matrix[idx])
train_index.append(df[df[label]==k].iloc[idx][id_column])
train_DF=pd.DataFrame(train_matrix,columns=columns_list)
test_DF=pd.DataFrame(test_matrix,columns=columns_list)
train_DF.index=np.array(train_index)
test_DF.index=np.array(test_index)
if(handle_unbalance):
train_DF=handle_unbalanced_dataset(train_DF,numeric_features,label,id_column)
return train_DF[numeric_features],test_DF[numeric_features],train_DF[label],test_DF[label]
def DT_RF_models(dataSet,numeric_features,path,isDT = True,iteration=10,testSize =0.2,readList = ['Compound Name'], label = 'Category',DTdenotion='test',DT_maxdepth=2,numberOfTrees = 50,RF_maxdepth=6,isplot=False,id_column='id',handle_unbalance=True):
if(isDT):
model=tree.DecisionTreeClassifier(max_depth=DT_maxdepth)
else:
model=RandomForestClassifier(n_estimators=numberOfTrees,max_depth=RF_maxdepth)
readableDF = dataSet.copy()
X = readableDF[numeric_features]
Y = readableDF[label]
readableDF[id_column]=readableDF[id_column].astype(str)
accuracy = []
fullWrongList=[]
fullTest=np.array([])
fullPredict=[]
for j in range(0,iteration):
X_train, X_test, Y_train, Y_test = cross_validation_split_with_unbalance_data(readableDF,numeric_features,label=label,id_column=id_column,test_size=testSize,handle_unbalance=handle_unbalance)
model = model.fit(X_train,Y_train)
pre_Y = model.predict(X_test)
pre_Y_pro= model.predict_proba(X_test)
Y_test = pd.DataFrame(Y_test)
Y_test[id_column]=X_test.index
# Y_test['index']=np.array([i for i in range(0,Y_test.shape[0])])
# Only for RF
if(not isDT):
for i in range(0,numberOfTrees):
single_tree = model.estimators_[i]
export_graphviz(single_tree,out_file=path+str(j)+'---tree---'+str(i)+'.dot', feature_names = X.columns,rounded = True, precision = 1)
if(isplot):
(graph, ) = pydot.graph_from_dot_file(path+str(j)+'---tree---'+str(i)+'.dot')
graph.write_png(path+str(j)+'---tree---'+str(i)+'.png')
count=0
for i in range(0,len(pre_Y)):
fullTest=np.append(fullTest,Y_test.iloc[i][label])
fullPredict.append(pre_Y_pro[i])
if(pre_Y[i] != Y_test.iloc[i][label]):
count = count+1
string=''
for l in range(0,len(readList)):
string = string + str(readableDF[readableDF[id_column]==Y_test.iloc[i][id_column]][readList[l]].values[0])+'---'
best_preds = np.argmax(pre_Y_pro[i])
singleWrongList = [pre_Y[i],string+Y_test.iloc[i][label],best_preds,pre_Y_pro[i],str(j)]
fullWrongList.append(singleWrongList)
print('------------------accuracy = '+str(1-count/len(pre_Y))+'------------------')
accuracy.append(1-count/len(pre_Y))
#Only for DT, plot DT
if(isDT & isplot):
newData=handle_unbalanced_dataset(dataSet,numeric_features,label=label,id_column=id_column)
model = model.fit(newData[numeric_features],newData[label])
dot_data = tree.export_graphviz(model,out_file=None,feature_names=X.columns,class_names=dataSet.groupby([label],as_index=False).count()[label].tolist(),filled=True,rounded=True,special_characters=True)
graph = graphviz.Source(dot_data)
graph.render(DTdenotion,view=True)
print(np.array(accuracy).mean(),np.array(accuracy).std())
labelList = list(set(fullTest))
labelList.sort()
labelMap= {labelList[i]:i for i in range(len(labelList))}
newfullTest=[labelMap[fullTest[i]] for i in range(len(fullTest))]
return accuracy,fullWrongList,newfullTest,np.array(fullPredict),labelList
def print_full_wrong_list(full_wrong_list):
s = set()
for i in full_wrong_list:
strings = 'Pre-Label: '+i[0]+' Details: '+i[1]+' Probabilities: '+str(i[3])
s.add(strings)
for i in s:
print(i)
def generate_features_values_dict(file):
f=open(file)
text = f.readline()
edges_dict={}
values_dict={}
features_dict={}
while(text):
regex = re.match(r"(\d+)\ ->\ (\d+)", text)
if regex:
if regex.groups()[0] in edges_dict:
edges_dict[regex.groups()[0]].append(regex.groups()[1])
else:
edges_dict[regex.groups()[0]] = [regex.groups()[1]]
regex2 = re.match(r"(\d+)\ \[label=\".+\[(.+)\]\"\]", text)
if regex2:
values_dict[regex2.groups()[0]] = regex2.groups()[1].split(', ')
regex3 = re.match(r"(\d+)\ \[label=\"(?!gini)(.+)\ <=*", text)
if regex3:
features_dict[regex3.groups()[1]]=regex3.groups()[0]
# print(text)
text = f.readline()
features_values_dict={key:[ values_dict[edges_dict[value][0]],values_dict[edges_dict[value][1]] ] for (key,value) in features_dict.items() }
f.close()
return features_values_dict
def generate_RF_feature_importance(path,df,numeric_features,label):
dfc=df.copy()
categories=len(dfc[label].unique())
regex = re.compile(r"\ +", re.IGNORECASE)
files = [path+f for f in listdir(path) if f.endswith('.dot') if not f.startswith('.')]
all_features_dict = {feature:list(np.zeros(categories+1)) for feature in numeric_features}
for file in files:
features_values_dict = generate_features_values_dict(file)
for (key,value) in features_values_dict.items():
key = regex.sub(' ',key.strip(" "))
tempList=[]
count=0
for i in range(0,len(all_features_dict[key])-1):
tempList.append(all_features_dict[key][i]+int(value[1][i])-int(value[0][i]))
count=count+int(value[1][i])+int(value[0][i])
tempList.append(count+all_features_dict[key][len(all_features_dict[key])-1])
all_features_dict[key]=tempList
matrix = []
for (key,value) in all_features_dict.items():
abscount=0
list_temp=[key]
for i in range(0,len(value)-1):
abscount=abscount+abs(value[i])
for i in range(0,len(value)-1):
if(abscount>0):
list_temp.append(value[i]/abscount)
else:
list_temp.append(0)
list_temp.append(abscount)
matrix.append(list_temp)
DF = pd.DataFrame(matrix)
DF.columns = ['Features']+dfc.groupby([label],as_index=False).count()[label].tolist()+['Sample Size']
DF.fillna(0)
return DF
def transform_feature_importance(fullFeatureImportanceDF,label_list):
feature_importance_DF = fullFeatureImportanceDF.copy()
for i in label_list:
feature_importance_DF[i] = round(feature_importance_DF[i],3)
feature_importance_DF['abs_'+i] = abs(feature_importance_DF[i])
feature_importance_DF['max_value'] = feature_importance_DF[['abs_'+i for i in label_list]].T.max()
feature_importance_DF['median_value'] = feature_importance_DF[['abs_'+i for i in label_list]].T.median()
feature_importance_DF['sampleSize_value']=pow(feature_importance_DF['Sample Size'],0.25)
feature_importance_DF['Ability']=feature_importance_DF['max_value']*feature_importance_DF['median_value']*feature_importance_DF['sampleSize_value']*10+5
feature_importance_DF = feature_importance_DF.sort_values(by='Ability', ascending=False)
return feature_importance_DF[['Features']+label_list+['Sample Size','Ability']]
#####plot histogram based on a list of values#####
def plot_histogram(title, measured,outputFilePath, bins_number = 1000):
output_file(outputFilePath)
hist, edges = np.histogram(measured, density=True, bins=bins_number)
p = figure(title=title, plot_width = 750, plot_height = 750,tools='', background_fill_color="#fafafa")
p.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:],
fill_color="navy", line_color="white", alpha=0.5)
p.y_range.start = 0
p.legend.location = "center_right"
p.legend.background_fill_color = "#fefefe"
p.xaxis.axis_label = 'x'
p.yaxis.axis_label = 'Pr(x)'
p.grid.grid_line_color="white"
p.x_range = Range1d(0,1.01)
show(p)
return p
def plotBWScatter(DataFrame ,xvalue,yvalue, sizevalue, outputFilePath, readList,plotWidth = 1200, plotHeight = 900, titleName='features importance'):
hover = HoverTool()
tooltipString = ""
for ele in readList:
readTuple = (ele.lower(),ele)
tooltipString = tooltipString + """<br><font face="Arial" size="4">%s: @%s<font>""" % readTuple
hover.tooltips = tooltipString
tools= [hover,WheelZoomTool(),PanTool(),BoxZoomTool(),ResetTool(),SaveTool()]
source= ColumnDataSource(DataFrame)
p = figure(plot_width = plotWidth, plot_height = plotHeight, tools=tools,title=titleName,toolbar_location='right',x_axis_label=xvalue.lower(),y_axis_label=yvalue.lower(),background_fill_color='white',title_location = 'above')
p.title.text_font_size='15pt'
p.title.align = 'center'
p.xaxis.axis_label_text_font_size='12pt'
p.yaxis.axis_label_text_font_size='12pt'
p.x_range = Range1d(DataFrame[xvalue].min()*1.1,DataFrame[xvalue].max()*1.1)
p.y_range = Range1d(DataFrame[yvalue].min()*1.1,DataFrame[yvalue].max()*1.1)
p.circle(x = xvalue,y = yvalue,size=sizevalue,source=source,color='grey')
p.toolbar.active_scroll=p.select_one(WheelZoomTool)#set default active to scroll tool
if outputFilePath.endswith('png'):
export_png(p, filename=outputFilePath)
else:
output_file(outputFilePath)
show(p)
#k-means clustering method
def k_means_DF(data_frame,numeric_features,clusters=8,is_row=True):
clustering_data_validation = data_frame[numeric_features].copy()
if(is_row):
corr_validation_DF = clustering_data_validation.T.corr()
else:
corr_validation_DF = clustering_data_validation.corr()
kmeans = KMeans(n_clusters=clusters,random_state=100).fit(corr_validation_DF)
clusterDic = {corr_validation_DF.columns[i]:kmeans.labels_[i] for i in range(0,len(kmeans.labels_))}
npArray = np.array([[key,value] for (key,value) in clusterDic.items() ])
DF = pd.DataFrame(npArray)
DF.columns = ['element','group']
return DF
def plotHeatMap(corrDF , featureList,path_file):
output_file(path_file)
corrDF.columns.name = 'Features'
df = pd.DataFrame(corrDF[featureList].stack(), columns=['Distance']).reset_index()
df.columns=['level_0','Features','Distance']
source = ColumnDataSource(df)
colors = ["#75968f", "#a5bab7", "#c9d9d3", "#e2e2e2", "#dfccce", "#ddb7b1", "#cc7878", "#933b41", "#550b1d"]
mapper = LinearColorMapper(palette=colors, low=df.Distance.min(), high=df.Distance.max())
p = figure(plot_width=3500, plot_height=3500, title="HeatMap",
x_range=featureList, y_range=featureList,
toolbar_location=None, tools="", x_axis_location="above")
p.rect(x="Features", y="level_0", width=1, height=1, source=source,line_color=None, fill_color=transform('Distance', mapper))
color_bar = ColorBar(color_mapper=mapper, location=(0, 0),
ticker=BasicTicker(desired_num_ticks=len(colors)),
formatter=PrintfTickFormatter(format="%d%%"))
p.add_layout(color_bar, 'right')
p.axis.axis_line_color = None
p.axis.major_tick_line_color = None
p.axis.major_label_text_font_size = "30pt"
p.axis.major_label_standoff = 0
p.xaxis.major_label_orientation = 1.0
show(p)
def heatMap(DF , path_file, x_size=3500, y_size=3500,font_size="15pt"):
featureList=DF.columns.tolist()
indexList=DF.index.tolist()
DF.columns.name = 'Features'
df = pd.DataFrame(DF[featureList].stack(), columns=['Distance']).reset_index()
df.columns=['level_0','Features','Distance']
source = ColumnDataSource(df)
colors = ["#75968f", "#a5bab7", "#c9d9d3", "#e2e2e2", "#dfccce", "#ddb7b1", "#cc7878", "#933b41", "#550b1d"]
mapper = LinearColorMapper(palette=colors, low=df.Distance.min(), high=df.Distance.max())
p = figure(plot_width=x_size, plot_height=y_size, title="HeatMap",
x_range=featureList, y_range=indexList,
toolbar_location=None, tools="", x_axis_location="above")
p.rect(x="Features", y="level_0", width=1, height=1, source=source,line_color=None, fill_color=transform('Distance', mapper))
color_bar = ColorBar(color_mapper=mapper, location=(0, 0),
ticker=BasicTicker(desired_num_ticks=len(colors)),
formatter=PrintfTickFormatter(format="%.2f"))
p.add_layout(color_bar, 'right')
p.axis.axis_line_color = None
p.axis.major_tick_line_color = None
p.axis.major_label_standoff = 0
p.xaxis.major_label_orientation = 1.0
p.axis.major_label_text_font_size = font_size
if path_file.endswith('png'):
export_png(p, filename=path_file)
else:
output_file(path_file)
show(p)
def plot_heatmap_for_kmeans_groups(data_frame,numeric_features,path,clusters=8,is_row=True):
result_DF = k_means_DF(data_frame,numeric_features,clusters,is_row)
for k in range(0,clusters):
group_filter = result_DF['group'].astype(str)==str(k)
subFeatureList = result_DF[group_filter]['element'].values
if(is_row):
subNormalData = data_frame[numeric_features].T[subFeatureList].copy()
else:
subNormalData = data_frame[subFeatureList].copy()
if(subNormalData.shape[1]<2):
continue
subcorrDF = subNormalData.corr()
subcorrDF.columns=[str(i) for i in subcorrDF.columns.tolist()]
assert len(subFeatureList) == subcorrDF.shape[0]
subDistMatrix = subcorrDF.values
for i in range(0,len(subDistMatrix)):
subDistMatrix[i]=1-subDistMatrix[i]
subDistMatrix = ssd.squareform(subDistMatrix)
sublinked = linkage(subDistMatrix,'ward','euclidean',True)
subFeatureDict= {i:[subcorrDF.columns[i]] for i in range(0,len(subcorrDF.columns))}
for i in range(0,len(sublinked)):
index = i+sublinked.shape[0]+1
firstList = subFeatureDict[sublinked[i][0]]
for j in subFeatureDict[sublinked[i][1]]:
firstList.append(j)
if(len(firstList)!=sublinked[i][3]):
print("the length is not equal")
subFeatureDict[index]=firstList
subFeatureList=subFeatureDict[sublinked.shape[0]*2]
strFeatureList = [str(i) for i in subFeatureList]
subcorrDF.index=subcorrDF.columns
subcorrDF=subcorrDF.T[subFeatureList].T
plotHeatMap(subcorrDF[subFeatureList].reset_index(drop=True),strFeatureList,path+'/heatmap-'+str(k)+'.html')
if clusters==1:
return strFeatureList
def plot_precision_recall_curve(full_test,full_predict,label_list,class_num=4,title='ROC curve'):
if(class_num==2):
full_test=label_binarize(full_test,classes=list(range(0,3)))
full_test=np.array([np.array([i[0],i[1]]) for i in full_test])
else:
full_test=label_binarize(full_test,classes=list(range(0,class_num)))
precision = dict()
recall = dict()
average_precision=dict()
for i in range(0,class_num):
precision[i],recall[i],_ = precision_recall_curve(full_test[:,i],full_predict[:,i])
average_precision[i] = average_precision_score(full_test[:,i],full_predict[:,i])
precision['micro'],recall['micro'],_=precision_recall_curve(full_test.ravel(),full_predict.ravel())
colors = cycle(['navy', 'turquoise', 'darkorange', 'cornflowerblue', 'teal'])
plt.figure(figsize=(7, 8))
labels = []
lines = []
for i, color in zip(range(class_num), colors):
l, = plt.plot(recall[i], precision[i], color=color, lw=2)
lines.append(l)
labels.append('Precision-recall for class {0} (AUC = {1:0.2f})'
''.format(label_list[i], average_precision[i]))
fig = plt.gcf()
fig.subplots_adjust(bottom=0.25)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('Recall')
plt.ylabel('Precision')
plt.title(title)
plt.legend(lines, labels, loc=(0, -.38), prop=dict(size=14))
plt.show()
def print_precision_recall_accuracy(full_test,full_predict,label_list,class_num=4):
right=0
wrong=0
for i in range(len(full_test)):
if(np.argmax(full_predict[i]) == int(full_test[i])):
right =right+1
else:
wrong=wrong+1
print("Overall Accuray: ",right/(right+wrong))
for n in range(class_num):
tp=0
fp=0
fn=0
for i in range(len(full_test)):
if(np.argmax(full_predict[i])==n):
if(np.argmax(full_predict[i]) == int(full_test[i])):
tp=tp+1
else:
fp=fp+1
elif(int(full_test[i])==n):
fn=fn+1
print(label_list[n],"label size:",tp+fn)
print(label_list[n],"Recall: ",tp/(tp+fn))
if((tp+fp)==0):
print(label_list[n],"Precision: ",0)
else:
print(label_list[n],"Precision: ",tp/(tp+fp))
##########Venn-Abers Predictor##########
### This part of codes is taken from https://github.com/ptocca/VennABERS, All credit of this part goes to the author of this repository.###
# Some elementary functions to speak the same language as the paper
# (at some point we'll just replace the occurrence of the calls with the function body itself)
def push(x,stack):
stack.append(x)
def pop(stack):
return stack.pop()
def top(stack):
return stack[-1]
def nextToTop(stack):
return stack[-2]
# perhaps inefficient but clear implementation
def nonleftTurn(a,b,c):
d1 = b-a
d2 = c-b
return np.cross(d1,d2)<=0
def nonrightTurn(a,b,c):
d1 = b-a
d2 = c-b
return np.cross(d1,d2)>=0
def slope(a,b):
ax,ay = a
bx,by = b
return (by-ay)/(bx-ax)
def notBelow(t,p1,p2):
p1x,p1y = p1
p2x,p2y = p2
tx,ty = t
m = (p2y-p1y)/(p2x-p1x)
b = (p2x*p1y - p1x*p2y)/(p2x-p1x)
return (ty >= tx*m+b)
kPrime = None
# Because we cannot have negative indices in Python (they have another meaning), I use a dictionary
def algorithm1(P):
global kPrime
S = []
P[-1] = np.array((-1,-1))
push(P[-1],S)
push(P[0],S)
#put P[0] at the end of S
for i in range(1,kPrime+1):
#nextToTop(S):S[len(S)-2] top(S):S[len(S)-1] pop(S):drop the last element
#cross product for 2 dimension vector return the value of axis z
#cross product vector of vec1 and vec2 is the perpendicular vector with the plane consist by vec1 and vec2
while len(S)>1 and nonleftTurn(nextToTop(S),top(S),P[i]):
pop(S)
push(P[i],S)
return S
def algorithm2(P,S):
global kPrime
Sprime = S[::-1] # reverse the stack
F1 = np.zeros((kPrime+1,))
for i in range(1,kPrime+1):
F1[i] = slope(top(Sprime),nextToTop(Sprime))
P[i-1] = P[i-2]+P[i]-P[i-1]
if notBelow(P[i-1],top(Sprime),nextToTop(Sprime)):
continue
pop(Sprime)
while len(Sprime)>1 and nonleftTurn(P[i-1],top(Sprime),nextToTop(Sprime)):
pop(Sprime)
push(P[i-1],Sprime)
return F1
def algorithm3(P):
global kPrime
P[kPrime+1] = P[kPrime]+np.array((1.0,0.0))
S = []
push(P[kPrime+1],S)
push(P[kPrime],S)
for i in range(kPrime-1,0-1,-1): # k'-1,k'-2,...,0
while len(S)>1 and nonrightTurn(nextToTop(S),top(S),P[i]):
pop(S)
push(P[i],S)
return S
def algorithm4(P,S):
global kPrime
Sprime = S[::-1] # reverse the stack
F0 = np.zeros((kPrime+1,))
for i in range(kPrime,1-1,-1): # k',k'-1,...,1
F0[i] = slope(top(Sprime),nextToTop(Sprime))
P[i] = P[i-1]+P[i+1]-P[i]
if notBelow(P[i],top(Sprime),nextToTop(Sprime)):
continue
pop(Sprime)
while len(Sprime)>1 and nonrightTurn(P[i],top(Sprime),nextToTop(Sprime)):
pop(Sprime)
push(P[i],Sprime)
return F0[1:]
def prepareData(calibrPoints):
global kPrime
#sort score_label_list based on ascending score
ptsSorted = sorted(calibrPoints)
#xs score np.array, ys, label np.array, both sorted
xs = np.fromiter((p[0] for p in ptsSorted),float)
ys = np.fromiter((p[1] for p in ptsSorted),float)
ptsUnique,ptsIndex,ptsInverse,ptsCounts = np.unique(xs,
return_index=True,
return_counts=True,
return_inverse=True)
a = np.zeros(ptsUnique.shape)
#a: for a unique score, how many items labeled 1.
np.add.at(a,ptsInverse,ys)
# now a contains the sums of ys for each unique value of the objects
w = ptsCounts
yPrime = a/w
#yPrime: the purity of label for each unique score
yCsd = np.cumsum(w*yPrime) # Might as well do just np.cumsum(a)
#yCsd accumulation of label1 through unique score list
xPrime = np.cumsum(w)
#xPrime: accumulation of observations through unique score list
kPrime = len(xPrime)
#kPrime: the number of unique scores
return yPrime,yCsd,xPrime,ptsUnique
def computeF(xPrime,yCsd):
P = {0:np.array((0,0))}
P.update({i+1:np.array((k,v)) for i,(k,v) in enumerate(zip(xPrime,yCsd))})
#P is (i->(xPrime[i],yCsd[i]))
S = algorithm1(P)
F1 = algorithm2(P,S)
# P = {}
# P.update({i+1:np.array((k,v)) for i,(k,v) in enumerate(zip(xPrime,yCsd))})
S = algorithm3(P)
F0 = algorithm4(P,S)
return F0,F1
def getFVal(F0,F1,ptsUnique,testObjects):
pos0 = np.searchsorted(ptsUnique[1:],testObjects,side='right')
pos1 = np.searchsorted(ptsUnique[:-1],testObjects,side='left')+1
return F0[pos0],F1[pos1]
def ScoresToMultiProbs(calibrPoints,testObjects):
# sort the points, transform into unique objects, with weights and updated values
yPrime,yCsd,xPrime,ptsUnique = prepareData(calibrPoints)
# compute the F0 and F1 functions from the CSD
F0,F1 = computeF(xPrime,yCsd)
# compute the values for the given test objects
p0,p1 = getFVal(F0,F1,ptsUnique,testObjects)
return p0,p1
def computeF1(yCsd,xPrime):
global kPrime
P = {0:np.array((0,0))}
P.update({i+1:np.array((k,v)) for i,(k,v) in enumerate(zip(xPrime,yCsd))})
S = algorithm1(P)
F1 = algorithm2(P,S)
return F1
def ScoresToMultiProbsV2(calibrPoints,testObjects):
# sort the points, transform into unique objects, with weights and updated values
yPrime,yCsd,xPrime,ptsUnique = prepareData(calibrPoints)
# compute the F0 and F1 functions from the CSD
F1 = computeF1(yCsd,xPrime)
pos1 = np.searchsorted(ptsUnique[:-1],testObjects,side='left')+1
p1 = F1[pos1]
yPrime,yCsd,xPrime,ptsUnique = prepareData((-x,1-y) for x,y in calibrPoints)
F0 = 1 - computeF1(yCsd,xPrime)
pos0 = np.searchsorted(ptsUnique[:-1],testObjects,side='left')+1
p0 = F0[pos0]
return p0,p1
def generate_label_from_probability(p0,p1,testScores,isprint=True):
p = p1/(1-p0+p1)
full_test=np.array([np.array([1-i,i]) for i in p])
t_p=[(int(round(testScores[i])),int(round(p[i])),testScores[i],p[i]) for i in range(0,len(p))]
#label from score, label from probability, score, probability
count=0
for i in range(0,len(t_p)):
if (t_p[i][0]!=t_p[i][1]):
count = count+1
if(isprint):
print("differ",count,t_p[i])
return t_p,full_test
##########End of Venn-Abers Predictor##########
def xgboostModel_for_venn(train,test,selectedData_Indices,n_vap=10,label = 'Control',category = 'Category',num_round = 100):
train_temp=train.copy()
test_temp=test.copy()
train_temp['cv_id']=train_temp.index
labelList = [label,'ZZZZZZZ']
train_temp.loc[train_temp[category]!=label,category]='ZZZZZZZ'
test_temp.loc[test_temp[category]!=label,category]='ZZZZZZZ'
score_and_label_list=[]
test_score=[]
regex = re.compile(r"\[|\]|<|\ ", re.IGNORECASE)
param = {'max_depth':2,'eta':0.3,'silent':1,'objective':'binary:logistic','learningrate':0.1} #'binary:logistic' 'multi:softprob' 'num_class':2,
train_temp.columns = [regex.sub('_',col) for col in train_temp.columns.values]
test_temp.columns = [regex.sub('_',col) for col in test_temp.columns.values]
selectedData_Indices = [regex.sub('_',col) for col in selectedData_Indices]
labelEncoder = LabelEncoder()
for _ in range(n_vap):
X_train,X_test,Y_train,Y_test = cross_validation_split_with_unbalance_data(train_temp.reset_index(drop=True).copy(),selectedData_Indices,label=category,id_column='cv_id',test_size=0.2,handle_unbalance=False)
Y_train = labelEncoder.fit_transform(Y_train.values)
Y_test = labelEncoder.fit_transform(Y_test.values)
dtrain = xgb.DMatrix(X_train,label=Y_train)
dtest = xgb.DMatrix(X_test,label=Y_test)
bst = xgb.train(param,dtrain,num_round,feval='map5eval',maximize=True)
preds = bst.predict(dtest)
best_preds = np.asarray([round(value) for value in preds])
Y_test = pd.DataFrame(Y_test).reset_index()
for i in range(0,len(best_preds)):
score_and_label_list.append((preds[i],Y_test.iloc[i][0]))
count=0
X_train=train_temp[selectedData_Indices]
X_test=test_temp[selectedData_Indices]
Y_train=train_temp[category]
Y_test=test_temp[category]
Y_train = labelEncoder.fit_transform(Y_train.values)
Y_test = labelEncoder.fit_transform(Y_test.values)
dtrain = xgb.DMatrix(X_train,label=Y_train)
dtest = xgb.DMatrix(X_test,label=Y_test)
bst = xgb.train(param,dtrain,num_round,feval='map5eval',maximize=True)
preds = bst.predict(dtest)
best_preds = np.asarray([round(value) for value in preds])
Y_test = pd.DataFrame(Y_test).reset_index()
for i in range(0,len(best_preds)):
test_score.append(preds[i])
if(best_preds[i] != Y_test.iloc[i][0]):
count=count+1
fullPredict=[np.array([1-i,i]) for i in list(preds)]
p0,p1 = ScoresToMultiProbs(score_and_label_list,test_score)
label_from_probability,full_predic_venn = generate_label_from_probability(p0,p1,test_score,False)
readable_pre=[i[0] for i in full_predic_venn]
test[label]=readable_pre
return test,Y_test[0].to_numpy(),np.array(fullPredict),labelList
def tSNEPlot(oriData,data_Indices,read_list,color_col,storing_loc,size_col = 5, iters=1000, perp=2, title='tSNE',num_components=2):
tsne = TSNE(n_components=num_components,random_state=0,n_iter=iters,perplexity=perp)
tSNE_DF = oriData.copy()
tSNE_DF=tSNE_DF.reset_index(drop=True)
tSNE_DF_2d = (tSNE_DF[data_Indices] - tSNE_DF[data_Indices].mean()) / (tSNE_DF[data_Indices].max() - tSNE_DF[data_Indices].min()).fillna(0)
tSNE_DF_2d = tsne.fit_transform(tSNE_DF_2d.to_numpy())
tSNE_DF_2d = pd.DataFrame(tSNE_DF_2d).reset_index(drop=True)
tSNE_DF_2d.columns=[str(i) for i in range(1,1+num_components)]
for i in read_list+[color_col]:
tSNE_DF_2d[i] = tSNE_DF[i]
tSNE_DF_2d[color_col]=tSNE_DF_2d[color_col].astype(str)
plotColorScatter(tSNE_DF_2d ,xvalue = '1',yvalue = '2', sizevalue = size_col, outputFilePath=storing_loc,plotWidth = 750, plotHeight = 750, readList = read_list,titleName=title,colorColumn=color_col,colorPattern=viridis)
return tSNE_DF_2d
def plotColorScatter(DataFrame ,xvalue = '0',yvalue = '1', sizevalue = 'size', outputFilePath='/abc/test.html',plotWidth = 750, plotHeight = 750, readList = ['1','2'],titleName='tSNE', colorColumn="Category", colorPattern=viridis):
factors = DataFrame[colorColumn].unique()
if len(factors)<8:
color_map = factor_cmap(colorColumn,factors=factors,palette=['#f03b20','#feb24c','#ffeda0','#636363','#a1d99b','#31a354','#3182bd'])
else:
color_map = factor_cmap(colorColumn,factors=factors,palette=colorPattern(len(factors)))
hover = HoverTool()
tooltipString = ""
for ele in readList:
ele=str(ele)
readTuple = (ele.lower(),ele)
tooltipString = tooltipString + """<br><font face="Arial" size="4">%s: @%s<font>""" % readTuple
hover.tooltips = tooltipString
tools= [hover,WheelZoomTool(),PanTool(),BoxZoomTool(),ResetTool(),SaveTool()]
source= ColumnDataSource(DataFrame)
output_file(outputFilePath)
p = figure(plot_width = plotWidth, plot_height = plotHeight, tools=tools,title=titleName,toolbar_location='right',x_axis_label=xvalue.lower(),y_axis_label=yvalue.lower(),background_fill_color='white',title_location = 'above')
p.title.text_font_size='15pt'
p.title.align = 'center'
p.xaxis.axis_label_text_font_size='12pt'
p.yaxis.axis_label_text_font_size='12pt'
p.x_range = Range1d(DataFrame[xvalue].min()*1.1,DataFrame[xvalue].max()*1.1)
p.y_range = Range1d(DataFrame[yvalue].min()*1.1,DataFrame[yvalue].max()*1.1)
p.circle(x = xvalue,y = yvalue,size=sizevalue,source=source,color=color_map,legend=colorColumn)
p.legend.location = "top_left"
p.toolbar.active_scroll=p.select_one(WheelZoomTool)
if outputFilePath.endswith('png'):
export_png(p, filename=outputFilePath)
else:
output_file(outputFilePath)
show(p)
def print_full_wrong_list(full_wrong_list):
s = set()
for i in full_wrong_list:
strings = 'Pre-Label: '+i[0]+' Original_data: '+i[1]+' Probability: '+str(i[3])
s.add(strings)
for i in s:
print(i)
def xgboost_multi_classification(input_df,numeric_features_validation,iteration=10,test_size=0.2,max_depth=2,num_trees=50,label_column='Category',id_column='PlateID',handle_unbalance=True,readList=['PlateID','Compound Name']):
XGBData = input_df.copy()
selectedData_Indices = numeric_features_validation.copy() # data_Indices
num_class=len(XGBData[label_column].unique().tolist())
regex = re.compile(r"\[|\]|<|\ ", re.IGNORECASE)
param = {'max_depth':max_depth,'eta':0.3,'silent':1,'objective':'multi:softprob','num_class':num_class,'learningrate':0.1}
num_round = num_trees
labelList = XGBData.groupby([label_column],as_index=False).mean()[label_column].tolist()
label = 0
accuracy = []
X = XGBData.reset_index()[selectedData_Indices]
Y = XGBData.reset_index()[label_column]
X.columns = [regex.sub('_',col) for col in X.columns.values]
XGBData.columns = [regex.sub('_',col) for col in XGBData.columns.values]
selectedData_Indices = [regex.sub('_',col) for col in selectedData_Indices]
labelEncoder = LabelEncoder()
labelEncoded = labelEncoder.fit_transform(XGBData.reset_index()[label_column].values)
fullWrongList=[]
fullTest=np.array([])
fullPredict=[]
for j in range(0,iteration):
X_train, X_test, Y_train, Y_test = cross_validation_split_with_unbalance_data(XGBData,selectedData_Indices,label=label_column,id_column=id_column,test_size=test_size,handle_unbalance=handle_unbalance)
Y_train = labelEncoder.fit_transform(Y_train.values)
Y_test = labelEncoder.fit_transform(Y_test.values)
fullTest=np.concatenate((fullTest,Y_test),axis=0)
dtrain = xgb.DMatrix(X_train,label=Y_train)
dtest = xgb.DMatrix(X_test,label=Y_test)
bst = xgb.train(param,dtrain,num_round,feval='map5eval',maximize=True)
preds = bst.predict(dtest)
fullPredict=fullPredict+list(preds)
best_preds = np.asarray([np.argmax(line) for line in preds])
precision = precision_score(Y_test,best_preds,average='macro')
Y_test = pd.DataFrame(Y_test).reset_index()
count=0
for i in range(0,len(best_preds)):
if(best_preds[i] != Y_test.iloc[i][label]):
count=count+1
string=''
for l in range(0,len(readList)):
string = string + str(XGBData.reset_index().iloc[X_test.reset_index(drop=True).index[i]][readList[l]])+'---'
singleWrongList = [labelList[best_preds[i]],string+labelList[Y_test.iloc[i][label]],str(j),preds[i]]
fullWrongList.append(singleWrongList)
print('------------------accuracy = '+str(1-count/len(best_preds))+'------------------')
accuracy.append(1-count/len(best_preds))
#bst.dump_model(storePath)
pArray = np.array(accuracy)
print(pArray.mean(),pArray.std())
return pArray,fullWrongList,fullTest,np.array(fullPredict),labelList
def combined_eXGBT_classifier(training_set,numeric_features_validation,testing_set,n_vap=10,label_column = 'Category',max_depth=2,num_trees=50):
num_class=len(training_set[label_column].unique().tolist())
df_te = testing_set.copy()
for i in training_set[label_column].unique().tolist():
df_te,full_test,full_predict,label_list =xgboostModel_for_venn(training_set,df_te,numeric_features_validation,n_vap,label =i,category = label_column,num_round = num_trees)
XGBData = training_set.copy()
selectedData_Indices = numeric_features_validation # data_Indices
regex = re.compile(r"\[|\]|<|\ ", re.IGNORECASE)
param = {'max_depth':max_depth,'eta':0.3,'silent':1,'objective':'multi:softprob','num_class':num_class,'learningrate':0.1}
labelList = XGBData.groupby([label_column],as_index=False).mean()[label_column].tolist()
X = XGBData.reset_index()[selectedData_Indices]
Y = XGBData.reset_index()[label_column]
Z = df_te[selectedData_Indices]
X.columns = [regex.sub('_',col) for col in X.columns.values]
Z.columns = [regex.sub('_',col) for col in Z.columns.values]
labelEncoder = LabelEncoder()
labelEncoded = labelEncoder.fit_transform(Y.values)
dtrain = xgb.DMatrix(X,label=labelEncoded)
bst = xgb.train(param,dtrain,num_trees,feval='map5eval',maximize=True)
dtest = xgb.DMatrix(Z)
preds = bst.predict(dtest)
best_preds = np.asarray([np.argmax(line) for line in preds])
readable_pre=[labelList[i] for i in best_preds]
df_te['multi_eXGBT_pre_label']=readable_pre
return df_te
def transform_predict_result_DF(predict_result_DF,label_col,threshold=0.1):
id_col='predict_result_DF_indices'
predict_result_DF[id_col]=predict_result_DF.index
#This is for specific purposes.
try:
label_list = predict_result_DF[label_col].unique().tolist()
predict_result_DF['max']=predict_result_DF[label_list].T.max()
except KeyError as keyE:
label_list = predict_result_DF['multi_eXGBT_pre_label'].unique().tolist()
predict_result_DF['max']=predict_result_DF[label_list].T.max()
print('Notice: The predicted labels were used instead of full labels')
min_Filter = predict_result_DF['max']<threshold
predict_result_DF.loc[min_Filter,'F_label']=predict_result_DF.loc[min_Filter,'multi_eXGBT_pre_label']
max_Filter = predict_result_DF['max']>=threshold
for i in label_list:
analogue_filter = predict_result_DF['max']==predict_result_DF[i]
predict_result_DF.loc[analogue_filter&max_Filter,'F_label']=i
predict_result_DF = predict_result_DF.rename({'max': 'probability'}, axis='columns')
temp1= predict_result_DF.groupby([id_col,'F_label'], as_index=False).mean()[[id_col,'F_label','probability']]
temp1['ID']=temp1[id_col].astype(str)+temp1['probability'].astype(str)
temp2= temp1.groupby([id_col], as_index=False).max()[[id_col,'probability']]
temp2['ID']=temp2[id_col].astype(str)+temp2['probability'].astype(str)
temp3=temp2.merge(temp1, on='ID', how='left')[[id_col+'_x','probability_x','F_label']]
temp3.columns=[id_col,'confidence','predicted_label']
temp3.groupby(['predicted_label'], as_index=False).count()[['predicted_label','confidence']]
temp3=temp3.merge(predict_result_DF, on=id_col, how='left')[[id_col,'confidence','predicted_label',label_col]]
fake_filter = temp3[id_col].astype(str).str.startswith('fake')
return predict_result_DF,temp3[~fake_filter]
def generate_expressed_matrix(gene_dfc,select_gene ,group_col = 'group',id_col='cell'):
np_temp = gene_dfc[select_gene].to_numpy()
lst=[]
for i in np_temp:
lst_t=[]
for j in i:
if j==0:
lst_t.append(0)
else:
lst_t.append(1)
lst.append(lst_t)
lst=np.array(lst)
zero_test_df = pd.DataFrame(lst,columns=select_gene)
zero_test_df[[id_col,group_col]]=gene_dfc[[id_col,group_col]]
return zero_test_df
def Drode_DE_gene_detection(gene_dfc,select_gene,feature='gene',id_col='cell',group_col='group',is_unify=True):
if(is_unify):
gene_dfc_unify,_ = unify_df(gene_dfc,select_gene,id_col,[group_col])
else:
gene_dfc_unify=gene_dfc.copy()
gene_dfc_unify= gene_dfc_unify.reset_index(drop=True)
zero_test_df = generate_expressed_matrix(gene_dfc = gene_dfc_unify,select_gene = select_gene,group_col = group_col,id_col=id_col)
differentiated_df = pd.DataFrame()
for gene in select_gene:
print(gene)
temp = zero_test_df.groupby(by=[group_col,gene],as_index=False)[[id_col]].count().copy()
temp = pd.pivot_table(temp, index=group_col, columns=gene, values=id_col)
if zero_test_df[zero_test_df[gene]==0].shape[0]==0:
temp[0]=np.zeros(temp.shape[0])
temp['non_0_rate']=np.ones(temp.shape[0])
elif zero_test_df[zero_test_df[gene]==1].shape[0]==0:
temp[1]=np.zeros(temp.shape[0])
temp['non_0_rate']=np.zeros(temp.shape[0])
else:
temp=temp.fillna(0)
temp['non_0_rate']=temp[1]/(temp[0]+temp[1])
temp['size']=temp[0]+temp[1]
temp[group_col]=temp.index
temp_np = temp.to_numpy()
lst=[]
for i in range(len(temp_np)):
for j in range(i+1,len(temp_np)):
x = gene_dfc_unify[(gene_dfc_unify[group_col]==str(temp_np[i][4])) & (gene_dfc_unify[gene]!=0)][gene].to_numpy()
y = gene_dfc_unify[(gene_dfc_unify[group_col]==str(temp_np[j][4])) & (gene_dfc_unify[gene]!=0)][gene].to_numpy()
true_mean_x = gene_dfc[(gene_dfc[group_col]==str(temp_np[i][4])) & (gene_dfc[gene]!=0)][gene].to_numpy()
true_mean_y = gene_dfc[(gene_dfc[group_col]==str(temp_np[j][4])) & (gene_dfc[gene]!=0)][gene].to_numpy()
if((len(x)<3) & (len(y)<3)):
continue
elif len(x)<3:
mean_x=0
mean_y=y.mean()
same_distribution_pval = 1
elif len(y)<3:
mean_x=x.mean()
mean_y=0
same_distribution_pval = 1
else:
mean_x=x.mean()
mean_y=y.mean()
same_distribution_pval = (ks_2samp(x, y)[1])
if(len(true_mean_x)>0):
true_mean_x=true_mean_x.mean()
else:
true_mean_x=0
if(len(true_mean_y)>0):
true_mean_y=true_mean_y.mean()
else:
true_mean_y=0
if temp_np[i][0]+temp_np[j][0]==0: #all zeros
same_0rate_pval=1
elif temp_np[i][0]+temp_np[j][0]==temp_np[i][3]+temp_np[j][3]: #all ones
same_0rate_pval=1
else:
zscore, same_0rate_pval = sm.stats.proportions_ztest([temp_np[i][0], temp_np[j][0]], [temp_np[i][3], temp_np[j][3]], alternative='two-sided')
if same_0rate_pval<=0.05:
differentiated_pval=1-same_0rate_pval
elif same_distribution_pval<=0.05:
differentiated_pval=0.95-same_distribution_pval
else:
differentiated_pval=0.9-same_0rate_pval*same_distribution_pval
lst.append([gene,temp_np[i][4],temp_np[j][4],differentiated_pval,1-same_0rate_pval,1-same_distribution_pval,temp_np[i][2],temp_np[j][2],temp_np[i][1],temp_np[j][1],true_mean_x,true_mean_y])
temp_df = pd.DataFrame(lst,columns=[feature,'group_1','group_2','deprob','d0prob','d1prob','posrate_1','posrate_2','pos_1','pos_2','posmean_1','posmean_2'])
differentiated_df=differentiated_df.append(temp_df)
return differentiated_df
def select_de_gene(drode_de_gene_df,score_col='deprob',feature='gene',num=20):
drode_de_gene_df = drode_de_gene_df[drode_de_gene_df[score_col]>0.9].copy()
drode_de_gene_df['group']=drode_de_gene_df['group_1']+'***'+drode_de_gene_df['group_2']
groups=drode_de_gene_df['group'].unique().tolist()
group_df=pd.DataFrame()
for group in groups:
temp_df = drode_de_gene_df[drode_de_gene_df['group']==group][['gene',score_col,'group']].copy()
temp_df = temp_df.sort_values(by=score_col,ascending=False).reset_index(drop=True)
group_df = group_df.append(temp_df.iloc[0:num])
de_gene_dWT4 = group_df[feature].unique().tolist()
return de_gene_dWT4
def cluster_cells(gene_dfc,select_gene,group='CD4SP',group_col='group',id_col='cell',path='/home/ivan/Desktop/Project2/MyData/pipeline'):
cluster_df = gene_dfc[gene_dfc[group_col]==group][select_gene+[id_col,group_col]]
cluster_df = cluster_df.reset_index(drop=True)
cluster_df = rrd(cluster_df,id_col,True,False)
cluster_df = cluster_df.replace(0,1e-7)
mx=cluster_df[select_gene].to_numpy()
scaled_matrix = generate_unit_modules(mx, isrow=True, is_scale=False, simple_scale=True)
dis_mx=[]
for i in range(len(scaled_matrix)):
lst=[]
for j in range(len(scaled_matrix)):
lst.append(entropy(scaled_matrix[i], scaled_matrix[j], base=None))
dis_mx.append(lst)
dis_mx = np.array(dis_mx)
dis_df = pd.DataFrame(dis_mx,columns=cluster_df[id_col].tolist())
dis_df.index = cluster_df[id_col].tolist()
lst = plot_heatmap_for_kmeans_groups(data_frame=dis_df,numeric_features=cluster_df[id_col].tolist(),path=path, clusters=1, is_row=True)
return lst
def de_gene_description(drode_de_gene_df):
drode_de_gene_df_2=drode_de_gene_df.copy()
drode_de_gene_df_2['group_1']=drode_de_gene_df['group_2']
drode_de_gene_df_2['group_2']=drode_de_gene_df['group_1']
drode_de_gene_df_2['posmean_1']=drode_de_gene_df['posmean_2']
drode_de_gene_df_2['posmean_2']=drode_de_gene_df['posmean_1']
drode_de_gene_df_2['posrate_1']=drode_de_gene_df['posrate_2']
drode_de_gene_df_2['posrate_2']=drode_de_gene_df['posrate_1']
drode_de_gene_df_2=drode_de_gene_df_2.append(drode_de_gene_df)
cate_lst = drode_de_gene_df_2['group_1'].unique().tolist()
temp_mean_df = pd.pivot_table(drode_de_gene_df_2, index='gene', columns='group_2', values='posmean_2')
temp_mean_df = temp_mean_df[cate_lst]
temp_mean_df['name']=temp_mean_df.index+'_PELs'
temp_mean_df.index=temp_mean_df['name']
temp_mean_df=temp_mean_df.reset_index(drop=True)
temp_mean_df = temp_mean_df.T
temp_mean_df.columns = temp_mean_df.loc['name']
temp_mean_df = temp_mean_df.loc[cate_lst]
temp_mean_df.index = cate_lst
temp_rate_df= pd.pivot_table(drode_de_gene_df_2, index='gene', columns='group_2', values='posrate_2')
temp_rate_df = temp_rate_df[cate_lst]
temp_rate_df['name']=temp_rate_df.index+'_1-RUEs'
temp_rate_df.index=temp_rate_df['name']
temp_rate_df=temp_rate_df.reset_index(drop=True)
temp_rate_df = temp_rate_df.T
temp_rate_df.columns = temp_rate_df.loc['name']
temp_rate_df = temp_rate_df.loc[cate_lst]
temp_rate_df.index = cate_lst
return temp_mean_df,temp_rate_df
def plot_line_charts(df1,df2=None,width=1200,height=900,titleName='title',xlabel='subpopulations',ylabel='expression_level',ylabel_sec='expression_rate',outputFilePath = "/home/ivan/Desktop/Project2/twin_axis.html"):
df = df1.copy()
tools= [WheelZoomTool(),PanTool(),BoxZoomTool(),ResetTool(),SaveTool()]
p = figure(x_range=df.index.tolist(),plot_width = width, plot_height = height, \
tools=tools,title=titleName,toolbar_location='right',x_axis_label=xlabel,\
y_axis_label=ylabel,background_fill_color='white',title_location = 'above')
if len(df.columns.tolist())<6:
palette=['#f03b20','#feb24c','#3182bd','#636363','#31a354']
else:
palette=viridis(len(df.columns.tolist()))
p.y_range = Range1d(min(0,df.values.min()-abs(df.values.min()*0.05)), df.values.max()+abs(df.values.max()*0.05))
col_lst=df.columns.tolist()
df['index']=df.index
for i in range(0,len(col_lst)):
col = col_lst[i]
p.line(df.index, df[col], legend=col, line_width=3, color=palette[i])
p.circle(df.index, df[col], size=7,color=palette[i])
if df2 is not None:
df = df2.copy()
y_column2_range = 'expression_rate' + "_range"
p.extra_y_ranges = {y_column2_range: Range1d(start=min(0,df.values.min()-abs(df.values.min()*0.05)),end=df.values.max()+abs(df.values.max()*0.05))}
p.add_layout(LinearAxis(y_range_name=y_column2_range,axis_label=ylabel_sec), "right")
col_lst=df.columns.tolist()
df['index']=df.index
source= ColumnDataSource(df)
for i in range(0,len(col_lst)):
col = col_lst[i]
p.line(df.index,df[col],legend=col,line_width=3,y_range_name=y_column2_range,color=palette[i],line_dash="dashed")
p.circle('index', col, size=7,color=palette[i],y_range_name=y_column2_range,source=source)
p.legend.location = "top_left"
p.toolbar.active_scroll=p.select_one(WheelZoomTool)
p.title.text_font_size='15pt'
p.title.align = 'center'
p.xaxis.axis_label_text_font_size='12pt'
p.yaxis.axis_label_text_font_size='12pt'
if outputFilePath.endswith('png'):
export_png(p, filename=outputFilePath)
else:
output_file(outputFilePath)
show(p)
def h_bar_plot(bar_df,cat_col,num_col,sort_col,bottom_up=False,height=600,title='test',file_path='test.html'):
bar_df = bar_df.sort_values(by=sort_col,ascending=bottom_up)
cate=bar_df[cat_col].tolist()
num=bar_df[num_col].tolist()
p = figure(y_range=cate, plot_height=height, x_range=(min(num)-abs(min(num)*0.1), max(num)+abs(max(num)*0.1)), title=title,toolbar_location='right')
source = ColumnDataSource(data=dict(num=num, cate=cate))
color_map = LinearColorMapper(palette=viridis(len(set(num))), low=min(num), high=max(num))
p.hbar(y='cate', right='num', height=0.9,fill_color={'field': 'num', 'transform': color_map}, legend=None, source=source)
p.y_range.range_padding = 0.1
p.ygrid.grid_line_color = None
p.axis.minor_tick_line_color = None
p.outline_line_color = None
p.title.text_font_size='15pt'
p.title.align = 'center'
p.xaxis.axis_label_text_font_size='12pt'
p.yaxis.axis_label_text_font_size='12pt'
if file_path.endswith('png'):
export_png(p, filename=file_path)
else:
output_file(file_path)
show(p)
def pca_transformation(gene_df_full,drode_de_gene):
pca_df = gene_df_full.copy()
pca_df['indices']=pca_df.index
temp_mx = dtm(pca_df,drode_de_gene,sort_column='indices')
temp_mx = scale_matrix(temp_mx,isrow=False,simple_scale=True)
pca_df = mtd(temp_mx,drode_de_gene,pca_df,['group','indices'], sort_column='indices',asc=True)
pca_df.index = pca_df['indices']
del pca_df['indices']
pca_df['cell']=pca_df.index
pca = PCA(n_components=5,random_state=0)
principalComponents = pca.fit_transform(pca_df[drode_de_gene])
principalDf = | pd.DataFrame(data = principalComponents, columns = ['pc1', 'pc2','pc3','pc4','pc5']) | pandas.DataFrame |
import pandas as pd
import numpy as np
import math
import os
from scipy.interpolate import interp1d
import time
from sklearn.ensemble import RandomForestRegressor
import xgboost as xgb
from lightgbm import LGBMRegressor
from catboost import CatBoostRegressor
from information_measures import *
from joblib import Parallel, delayed
#from arch import arch_model
def rmspe(y_true, y_pred):
return (np.sqrt(np.mean(np.square((y_true - y_pred) / y_true))))
def log_return(list_stock_prices): # Stock prices are estimated through wap values
return np.log(list_stock_prices).diff()
def realized_volatility(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
def compute_wap(book_pd):
wap = (book_pd['bid_price1'] * book_pd['ask_size1'] + book_pd['ask_price1'] * book_pd['bid_size1']) / (book_pd['bid_size1']+ book_pd['ask_size1'])
return wap
def realized_volatility_from_book_pd(book_stock_time):
wap = compute_wap(book_stock_time)
returns = log_return(wap)
volatility = realized_volatility(returns)
return volatility
def realized_volatility_per_time_id(file_path, prediction_column_name):
df_book_data = pd.read_parquet(file_path)
# Estimate stock price per time point
df_book_data['wap'] = compute_wap(df_book_data)
# Compute log return from wap values per time_id
df_book_data['log_return'] = df_book_data.groupby(['time_id'])['wap'].apply(log_return)
df_book_data = df_book_data[~df_book_data['log_return'].isnull()]
# Compute the square root of the sum of log return squared to get realized volatility
df_realized_vol_per_stock = pd.DataFrame(df_book_data.groupby(['time_id'])['log_return'].agg(realized_volatility)).reset_index()
# Formatting
df_realized_vol_per_stock = df_realized_vol_per_stock.rename(columns = {'log_return':prediction_column_name})
stock_id = file_path.split('=')[1]
df_realized_vol_per_stock['row_id'] = df_realized_vol_per_stock['time_id'].apply(lambda x:f'{stock_id}-{x}')
return df_realized_vol_per_stock[['row_id',prediction_column_name]]
def past_realized_volatility_per_stock(list_file,prediction_column_name):
df_past_realized = pd.DataFrame()
for file in list_file:
df_past_realized = pd.concat([df_past_realized,
realized_volatility_per_time_id(file,prediction_column_name)])
return df_past_realized
def stupidForestPrediction(book_path_train,prediction_column_name,train_targets_pd,book_path_test):
naive_predictions_train = past_realized_volatility_per_stock(list_file=book_path_train,prediction_column_name=prediction_column_name)
df_joined_train = train_targets_pd.merge(naive_predictions_train[['row_id','pred']], on = ['row_id'], how = 'left')
X = np.array(df_joined_train['pred']).reshape(-1,1)
y = np.array(df_joined_train['target']).reshape(-1,)
regr = RandomForestRegressor(random_state=0)
regr.fit(X, y)
naive_predictions_test = past_realized_volatility_per_stock(list_file=book_path_test,prediction_column_name='target')
yhat = regr.predict(np.array(naive_predictions_test['target']).reshape(-1,1))
updated_predictions = naive_predictions_test.copy()
updated_predictions['target'] = yhat
return updated_predictions
def garch_fit_predict_volatility(returns_series, N=10000):
model = arch_model(returns_series * N, p=1, q=1)
model_fit = model.fit(update_freq=0, disp='off')
yhat = model_fit.forecast(horizon=600, reindex=False)
pred_volatility = np.sqrt(np.sum(yhat.variance.values)) / N
return pred_volatility
def garch_volatility_per_time_id(file_path, prediction_column_name):
# read the data
df_book_data = pd.read_parquet(file_path)
# calculate the midprice (not the WAP)
df_book_data['midprice'] =(df_book_data['bid_price1'] + df_book_data['ask_price1'])/2
# leave only WAP for now
df_book_data = df_book_data[['time_id', 'seconds_in_bucket', 'midprice']]
df_book_data = df_book_data.sort_values('seconds_in_bucket')
# make the book updates evenly spaced
df_book_data_evenly = pd.DataFrame({'time_id':np.repeat(df_book_data['time_id'].unique(), 600),
'second':np.tile(range(0,600), df_book_data['time_id'].nunique())})
df_book_data_evenly['second'] = df_book_data_evenly['second'].astype(np.int16)
df_book_data_evenly = df_book_data_evenly.sort_values('second')
df_book_data_evenly = pd.merge_asof(df_book_data_evenly,
df_book_data,
left_on='second',right_on='seconds_in_bucket',
by = 'time_id')
# Ordering for easier use
df_book_data_evenly = df_book_data_evenly[['time_id', 'second', 'midprice']]
df_book_data_evenly = df_book_data_evenly.sort_values(['time_id','second']).reset_index(drop=True)
# calculate log returns
df_book_data_evenly['log_return'] = df_book_data_evenly.groupby(['time_id'])['midprice'].apply(log_return)
df_book_data_evenly = df_book_data_evenly[~df_book_data_evenly['log_return'].isnull()]
# fit GARCH(1, 1) and predict the volatility of returns
df_garch_vol_per_stock = \
pd.DataFrame(df_book_data_evenly.groupby(['time_id'])['log_return'].agg(garch_fit_predict_volatility)).reset_index()
df_garch_vol_per_stock = df_garch_vol_per_stock.rename(columns = {'log_return':prediction_column_name})
# add row_id column to the data
stock_id = file_path.split('=')[1]
df_garch_vol_per_stock['row_id'] = df_garch_vol_per_stock['time_id'].apply(lambda x:f'{stock_id}-{x}')
# return the result
return df_garch_vol_per_stock[['row_id', prediction_column_name]]
def garch_volatility_per_stock(list_file, prediction_column_name):
df_garch_predicted = pd.DataFrame()
for file in list_file:
df_garch_predicted = pd.concat([df_garch_predicted,
garch_volatility_per_time_id(file, prediction_column_name)])
return df_garch_predicted
def entropy_from_book(book_stock_time,last_min):
if last_min < 10:
book_stock_time = book_stock_time[book_stock_time['seconds_in_bucket'] >= (600-last_min*60)]
if book_stock_time.empty == True or book_stock_time.shape[0] < 3:
return 0
wap = compute_wap(book_stock_time)
t_init = book_stock_time['seconds_in_bucket']
t_new = np.arange(np.min(t_init),np.max(t_init))
# Closest neighbour interpolation (no changes in wap between lines)
nearest = interp1d(t_init, wap, kind='nearest')
resampled_wap = nearest(t_new)
# Compute sample entropy
# sampleEntropy = nolds.sampen(resampled_wap)
sampleEntropy = sampen(resampled_wap)
return sampleEntropy
def entropy_from_wap(wap,seconds,last_seconds):
if last_seconds < 600:
idx = np.where(seconds >= last_seconds)[0]
if len(idx) < 3:
return 0
else:
wap = wap[idx]
seconds = seconds[idx]
# Closest neighbour interpolation (no changes in wap between lines)
t_new = np.arange(np.min(seconds),np.max(seconds))
nearest = interp1d(seconds, wap, kind='nearest')
resampled_wap = nearest(t_new)
# Compute sample entropy
# sampleEntropy = nolds.sampen(resampled_wap)
sampleEntropy = sampen(resampled_wap)
# sampleEntropy = ApEn_new(resampled_wap,3,0.001)
return sampleEntropy
def linearFit(book_stock_time, last_min):
if last_min < 10:
book_stock_time = book_stock_time[book_stock_time['seconds_in_bucket'] >= (600-last_min*60)]
if book_stock_time.empty == True or book_stock_time.shape[0] < 2:
return 0
wap = np.array(compute_wap(book_stock_time))
t_init = book_stock_time['seconds_in_bucket']
return (wap[-1] - wap[0])/(np.max(t_init) - np.min(t_init))
def wapStat(book_stock_time, last_min):
if last_min < 10:
book_stock_time = book_stock_time[book_stock_time['seconds_in_bucket'] >= (600-last_min*60)]
if book_stock_time.empty == True or book_stock_time.shape[0] < 2:
return 0
wap = compute_wap(book_stock_time)
t_init = book_stock_time['seconds_in_bucket']
t_new = np.arange(np.min(t_init),np.max(t_init))
# Closest neighbour interpolation (no changes in wap between lines)
nearest = interp1d(t_init, wap, kind='nearest')
resampled_wap = nearest(t_new)
return np.std(resampled_wap)
def entropy_Prediction(book_path_train,prediction_column_name,train_targets_pd,book_path_test,all_stocks_ids,test_file):
# Compute features
book_features_encoded_test = computeFeatures_1(book_path_test,'test',test_file,all_stocks_ids)
book_features_encoded_train = computeFeatures_1(book_path_train,'train',train_targets_pd,all_stocks_ids)
X = book_features_encoded_train.drop(['row_id','target','stock_id'],axis=1)
y = book_features_encoded_train['target']
# Modeling
catboost_default = CatBoostRegressor(verbose=0)
catboost_default.fit(X,y)
# Predict
X_test = book_features_encoded_test.drop(['row_id','stock_id'],axis=1)
yhat = catboost_default.predict(X_test)
# Formatting
yhat_pd = pd.DataFrame(yhat,columns=['target'])
predictions = pd.concat([test_file,yhat_pd],axis=1)
return predictions
def computeFeatures_1(book_path,prediction_column_name,train_targets_pd,all_stocks_ids):
book_all_features = pd.DataFrame()
encoder = np.eye(len(all_stocks_ids))
stocks_id_list, row_id_list = [], []
volatility_list, entropy2_list = [], []
linearFit_list, linearFit5_list, linearFit2_list = [], [], []
wap_std_list, wap_std5_list, wap_std2_list = [], [], []
for file in book_path:
start = time.time()
book_stock = pd.read_parquet(file)
stock_id = file.split('=')[1]
print('stock id computing = ' + str(stock_id))
stock_time_ids = book_stock['time_id'].unique()
for time_id in stock_time_ids:
# Access book data at this time + stock
book_stock_time = book_stock[book_stock['time_id'] == time_id]
# Create feature matrix
stocks_id_list.append(stock_id)
row_id_list.append(str(f'{stock_id}-{time_id}'))
volatility_list.append(realized_volatility_from_book_pd(book_stock_time=book_stock_time))
entropy2_list.append(entropy_from_book(book_stock_time=book_stock_time,last_min=2))
linearFit_list.append(linearFit(book_stock_time=book_stock_time,last_min=10))
linearFit5_list.append(linearFit(book_stock_time=book_stock_time,last_min=5))
linearFit2_list.append(linearFit(book_stock_time=book_stock_time,last_min=2))
wap_std_list.append(wapStat(book_stock_time=book_stock_time,last_min=10))
wap_std5_list.append(wapStat(book_stock_time=book_stock_time,last_min=5))
wap_std2_list.append(wapStat(book_stock_time=book_stock_time,last_min=2))
print('Computing one stock entropy took', time.time() - start, 'seconds for stock ', stock_id)
# Merge targets
stocks_id_pd = pd.DataFrame(stocks_id_list,columns=['stock_id'])
row_id_pd = pd.DataFrame(row_id_list,columns=['row_id'])
volatility_pd = pd.DataFrame(volatility_list,columns=['volatility'])
entropy2_pd = pd.DataFrame(entropy2_list,columns=['entropy2'])
linearFit_pd = pd.DataFrame(linearFit_list,columns=['linearFit_coef'])
linearFit5_pd = pd.DataFrame(linearFit5_list,columns=['linearFit_coef5'])
linearFit2_pd = pd.DataFrame(linearFit2_list,columns=['linearFit_coef2'])
wap_std_pd = pd.DataFrame(wap_std_list,columns=['wap_std'])
wap_std5_pd = pd.DataFrame(wap_std5_list,columns=['wap_std5'])
wap_std2_pd = pd.DataFrame(wap_std2_list,columns=['wap_std2'])
book_all_features = pd.concat([stocks_id_pd,row_id_pd,volatility_pd,entropy2_pd,linearFit_pd,linearFit5_pd,linearFit2_pd,
wap_std_pd,wap_std5_pd,wap_std2_pd],axis=1)
# This line makes sure the predictions are aligned with the row_id in the submission file
book_all_features = train_targets_pd.merge(book_all_features, on = ['row_id'])
# Add encoded stock
encoded = list()
for i in range(book_all_features.shape[0]):
stock_id = book_all_features['stock_id'][i]
encoded_stock = encoder[np.where(all_stocks_ids == int(stock_id))[0],:]
encoded.append(encoded_stock)
encoded_pd = pd.DataFrame(np.array(encoded).reshape(book_all_features.shape[0],np.array(all_stocks_ids).shape[0]))
book_all_features_encoded = pd.concat([book_all_features, encoded_pd],axis=1)
return book_all_features_encoded
def calc_wap(df):
return (df['bid_price1'] * df['ask_size1'] + df['ask_price1'] * df['bid_size1']) / (df['bid_size1'] + df['ask_size1'])
def calc_wap2(df):
return (df['bid_price2'] * df['ask_size2'] + df['ask_price2'] * df['bid_size2']) / (df['bid_size2'] + df['ask_size2'])
def calc_wap3(df):
return (df['bid_price2'] * df['bid_size2'] + df['ask_price2'] * df['ask_size2']) / (df['bid_size2'] + df['ask_size2'])
def calc_wap4(df):
return (df['bid_price1'] * df['bid_size1'] + df['ask_price1'] * df['ask_size1']) / (df['bid_size1'] + df['ask_size1'])
def mid_price(df):
return df['bid_price1'] /2 + df['ask_price1'] / 2
def calc_rv_from_wap_numba(values, index):
log_return = np.diff(np.log(values))
realized_vol = np.sqrt(np.sum(np.square(log_return[1:])))
return realized_vol
def load_book_data_by_id(stock_id,datapath,train_test):
file_to_read = os.path.join(datapath,'book_' + str(train_test) + str('.parquet'),'stock_id=' + str(stock_id))
df = pd.read_parquet(file_to_read)
return df
def load_trades_data_by_id(stock_id,datapath,train_test):
file_to_read = os.path.join(datapath,'trade_' + str(train_test) + str('.parquet'),'stock_id=' + str(stock_id))
df = pd.read_parquet(file_to_read)
return df
def entropy_from_df(df):
if df.shape[0] < 3:
return 0
t_init = df['seconds_in_bucket']
t_new = np.arange(np.min(t_init),np.max(t_init))
# Closest neighbour interpolation (no changes in wap between lines)
nearest = interp1d(t_init, df['wap'], kind='nearest')
resampled_wap = nearest(t_new)
# Compute sample entropy
# sampleEntropy = nolds.sampen(resampled_wap)
sampleEntropy = sampen(resampled_wap)
return sampleEntropy
def entropy_from_df2(df):
if df.shape[0] < 3:
return 0
t_init = df['seconds_in_bucket']
t_new = np.arange(np.min(t_init),np.max(t_init))
# Closest neighbour interpolation (no changes in wap between lines)
nearest = interp1d(t_init, df['wap2'], kind='nearest')
resampled_wap = nearest(t_new)
# Compute sample entropy
# sampleEntropy = nolds.sampen(resampled_wap)
sampleEntropy = sampen(resampled_wap)
return sampleEntropy
def entropy_from_df3(df):
if df.shape[0] < 3:
return 0
t_init = df['seconds_in_bucket']
t_new = np.arange(np.min(t_init),np.max(t_init))
# Closest neighbour interpolation (no changes in wap between lines)
nearest = interp1d(t_init, df['wap3'], kind='nearest')
resampled_wap = nearest(t_new)
# Compute sample entropy
sampleEntropy = sampen(resampled_wap)
return sampleEntropy
def financial_metrics(df):
wap_imbalance = np.mean(df['wap'] - df['wap2'])
price_spread = np.mean((df['ask_price1'] - df['bid_price1']) / ((df['ask_price1'] + df['bid_price1'])/2))
bid_spread = np.mean(df['bid_price1'] - df['bid_price2'])
ask_spread = np.mean(df['ask_price1'] - df['ask_price2']) # Abs to take
total_volume = np.mean((df['ask_size1'] + df['ask_size2']) + (df['bid_size1'] + df['bid_size2']))
volume_imbalance = np.mean(abs((df['ask_size1'] + df['ask_size2']) - (df['bid_size1'] + df['bid_size2'])))
return [wap_imbalance,price_spread,bid_spread,ask_spread,total_volume,volume_imbalance]
def financial_metrics_2(df):
wap_imbalance = df['wap'] - df['wap2']
price_spread = (df['ask_price1'] - df['bid_price1']) / ((df['ask_price1'] + df['bid_price1'])/2)
bid_spread = df['bid_price1'] - df['bid_price2']
ask_spread = df['ask_price1'] - df['ask_price2'] # Abs to take
total_volume = (df['ask_size1'] + df['ask_size2']) + (df['bid_size1'] + df['bid_size2'])
volume_imbalance = abs((df['ask_size1'] + df['ask_size2']) - (df['bid_size1'] + df['bid_size2']))
# New features here
wap_imbalance_mean = np.mean(wap_imbalance)
wap_imbalance_sum = np.sum(wap_imbalance)
wap_imbalance_std = np.std(wap_imbalance)
wap_imbalance_max = np.max(wap_imbalance)
wap_imbalance_min = np.min(wap_imbalance)
price_spread_mean = np.mean(price_spread)
price_spread_sum = np.sum(price_spread)
price_spread_std = np.std(price_spread)
price_spread_max = np.max(price_spread)
price_spread_min = np.min(price_spread)
bid_spread_mean = np.mean(bid_spread)
bid_spread_sum = np.sum(bid_spread)
bid_spread_std = np.std(bid_spread)
bid_spread_max = np.max(bid_spread)
bid_spread_min = np.min(bid_spread)
ask_spread_mean = np.mean(ask_spread)
ask_spread_sum = np.sum(ask_spread)
ask_spread_std = np.std(ask_spread)
ask_spread_max = np.max(ask_spread)
ask_spread_min = np.min(ask_spread)
total_volume_mean = np.mean(total_volume)
total_volume_sum = np.sum(total_volume)
total_volume_std = np.std(total_volume)
total_volume_max = np.max(total_volume)
total_volume_min = np.min(total_volume)
volume_imbalance_mean = np.mean(volume_imbalance)
volume_imbalance_sum = np.sum(volume_imbalance)
volume_imbalance_std = np.std(volume_imbalance)
volume_imbalance_max = np.max(volume_imbalance)
volume_imbalance_min = np.min(volume_imbalance)
return [wap_imbalance_mean,price_spread_mean,bid_spread_mean,ask_spread_mean,total_volume_mean,volume_imbalance_mean, wap_imbalance_sum,price_spread_sum,bid_spread_sum,ask_spread_sum,total_volume_sum,volume_imbalance_sum, wap_imbalance_std,price_spread_std,bid_spread_std,ask_spread_std,total_volume_std,volume_imbalance_std, wap_imbalance_max,price_spread_max,bid_spread_max,ask_spread_max,total_volume_max,volume_imbalance_max, wap_imbalance_min,price_spread_min,bid_spread_min,ask_spread_min,total_volume_min,volume_imbalance_min]
def other_metrics(df):
if df.shape[0] < 2:
linearFit = 0
linearFit2 = 0
linearFit3 = 0
std_1 = 0
std_2 = 0
std_3 = 0
else:
linearFit = (df['wap'].iloc[-1] - df['wap'].iloc[0]) / ((np.max(df['seconds_in_bucket']) - np.min(df['seconds_in_bucket'])))
linearFit2 = (df['wap2'].iloc[-1] - df['wap2'].iloc[0]) / ((np.max(df['seconds_in_bucket']) - np.min(df['seconds_in_bucket'])))
linearFit3 = (df['wap3'].iloc[-1] - df['wap3'].iloc[0]) / ((np.max(df['seconds_in_bucket']) - np.min(df['seconds_in_bucket'])))
# Resampling
t_init = df['seconds_in_bucket']
t_new = np.arange(np.min(t_init),np.max(t_init))
# Closest neighbour interpolation (no changes in wap between lines)
nearest = interp1d(t_init, df['wap'], kind='nearest')
nearest2 = interp1d(t_init, df['wap2'], kind='nearest')
nearest3 = interp1d(t_init, df['wap3'], kind='nearest')
std_1 = np.std(nearest(t_new))
std_2 = np.std(nearest2(t_new))
std_3 = np.std(nearest3(t_new))
return [linearFit, linearFit2, linearFit3, std_1, std_2, std_3]
def load_book_data_by_id_kaggle(stock_id,train_test):
df = pd.read_parquet(f'../input/optiver-realized-volatility-prediction/book_{train_test}.parquet/stock_id={stock_id}')
return df
def load_trades_data_by_id_kaggle(stock_id,train_test):
df = pd.read_parquet(f'../input/optiver-realized-volatility-prediction/trade_{train_test}.parquet/stock_id={stock_id}')
return df
def computeFeatures_wEntropy(machine, dataset, all_stocks_ids, datapath):
list_rv, list_rv2, list_rv3 = [], [], []
list_ent, list_fin, list_fin2 = [], [], []
list_others, list_others2, list_others3 = [], [], []
for stock_id in range(127):
start = time.time()
if machine == 'local':
try:
book_stock = load_book_data_by_id(stock_id,datapath,dataset)
except:
continue
elif machine == 'kaggle':
try:
book_stock = load_book_data_by_id_kaggle(stock_id,dataset)
except:
continue
# Useful
all_time_ids_byStock = book_stock['time_id'].unique()
# Calculate wap for the book
book_stock['wap'] = calc_wap(book_stock)
book_stock['wap2'] = calc_wap2(book_stock)
book_stock['wap3'] = calc_wap3(book_stock)
# Calculate realized volatility
df_sub = book_stock.groupby('time_id')['wap'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub2 = book_stock.groupby('time_id')['wap2'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub3 = book_stock.groupby('time_id')['wap3'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_sub['time_id']]
df_sub = pd.concat([df_sub,df_sub2['wap2'],df_sub3['wap3']],axis=1)
df_sub = df_sub.rename(columns={'time_id':'row_id','wap': 'rv', 'wap2': 'rv2', 'wap3': 'rv3'})
# Calculate realized volatility last 5 min
isEmpty = book_stock.query(f'seconds_in_bucket >= 300').empty
if isEmpty == False:
df_sub_5 = book_stock.query(f'seconds_in_bucket >= 300').groupby(['time_id'])['wap'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub2_5 = book_stock.query(f'seconds_in_bucket >= 300').groupby(['time_id'])['wap2'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub3_5 = book_stock.query(f'seconds_in_bucket >= 300').groupby(['time_id'])['wap3'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub_5['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_sub_5['time_id']]
df_sub_5 = pd.concat([df_sub_5,df_sub2_5['wap2'],df_sub3_5['wap3']],axis=1)
df_sub_5 = df_sub_5.rename(columns={'time_id':'row_id','wap': 'rv_5', 'wap2': 'rv2_5', 'wap3': 'rv3_5'})
else: # 0 volatility
times_pd = pd.DataFrame(all_time_ids_byStock,columns=['time_id'])
times_pd['time_id'] = [f'{stock_id}-{time_id}' for time_id in times_pd['time_id']]
times_pd = times_pd.rename(columns={'time_id':'row_id'})
zero_rv = pd.DataFrame(np.zeros((1,times_pd.shape[0])),columns=['rv_5'])
zero_rv2 = pd.DataFrame(np.zeros((1,times_pd.shape[0])),columns=['rv2_5'])
zero_rv3 = pd.DataFrame(np.zeros((1,times_pd.shape[0])),columns=['rv3_5'])
df_sub_5 = pd.concat([times_pd,zero_rv,zero_rv2,zero_rv3],axis=1)
# Calculate realized volatility last 2 min
isEmpty = book_stock.query(f'seconds_in_bucket >= 480').empty
if isEmpty == False:
df_sub_2 = book_stock.query(f'seconds_in_bucket >= 480').groupby(['time_id'])['wap'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub2_2 = book_stock.query(f'seconds_in_bucket >= 480').groupby(['time_id'])['wap2'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub3_2 = book_stock.query(f'seconds_in_bucket >= 480').groupby(['time_id'])['wap3'].agg(calc_rv_from_wap_numba, engine='numba').to_frame().reset_index()
df_sub_2['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_sub_2['time_id']]
df_sub_2 = pd.concat([df_sub_2,df_sub2_2['wap2'],df_sub3_2['wap3']],axis=1)
df_sub_2 = df_sub_2.rename(columns={'time_id':'row_id','wap': 'rv_2', 'wap2': 'rv2_2', 'wap3': 'rv3_2'})
else: # 0 volatility
times_pd = pd.DataFrame(all_time_ids_byStock,columns=['time_id'])
times_pd['time_id'] = [f'{stock_id}-{time_id}' for time_id in times_pd['time_id']]
times_pd = times_pd.rename(columns={'time_id':'row_id'})
zero_rv = pd.DataFrame(np.zeros((1,times_pd.shape[0])),columns=['rv_2'])
zero_rv2 = pd.DataFrame(np.zeros((1,times_pd.shape[0])),columns=['rv2_2'])
zero_rv3 = pd.DataFrame(np.zeros((1,times_pd.shape[0])),columns=['rv3_2'])
df_sub_2 = pd.concat([times_pd,zero_rv,zero_rv2,zero_rv3],axis=1)
list_rv.append(df_sub)
list_rv2.append(df_sub_5)
list_rv3.append(df_sub_2)
# Calculate other financial metrics from book
df_sub_book_feats = book_stock.groupby(['time_id']).apply(financial_metrics).to_frame().reset_index()
df_sub_book_feats = df_sub_book_feats.rename(columns={0:'embedding'})
df_sub_book_feats[['wap_imbalance','price_spread','bid_spread','ask_spread','total_vol','vol_imbalance']] = pd.DataFrame(df_sub_book_feats.embedding.tolist(), index=df_sub_book_feats.index)
df_sub_book_feats['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_sub_book_feats['time_id']]
df_sub_book_feats = df_sub_book_feats.rename(columns={'time_id':'row_id'}).drop(['embedding'],axis=1)
isEmpty = book_stock.query(f'seconds_in_bucket >= 300').empty
if isEmpty == False:
df_sub_book_feats5 = book_stock.query(f'seconds_in_bucket >= 300').groupby(['time_id']).apply(financial_metrics).to_frame().reset_index()
df_sub_book_feats5 = df_sub_book_feats5.rename(columns={0:'embedding'})
df_sub_book_feats5[['wap_imbalance5','price_spread5','bid_spread5','ask_spread5','total_vol5','vol_imbalance5']] = pd.DataFrame(df_sub_book_feats5.embedding.tolist(), index=df_sub_book_feats5.index)
df_sub_book_feats5['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_sub_book_feats5['time_id']]
df_sub_book_feats5 = df_sub_book_feats5.rename(columns={'time_id':'row_id'}).drop(['embedding'],axis=1)
else:
times_pd = pd.DataFrame(all_time_ids_byStock,columns=['time_id'])
times_pd['time_id'] = [f'{stock_id}-{time_id}' for time_id in times_pd['time_id']]
times_pd = times_pd.rename(columns={'time_id':'row_id'})
temp = pd.DataFrame([0],columns=['wap_imbalance5'])
temp2 = pd.DataFrame([0],columns=['price_spread5'])
temp3 = pd.DataFrame([0],columns=['bid_spread5'])
temp4 = pd.DataFrame([0],columns=['ask_spread5'])
temp5 = pd.DataFrame([0],columns=['total_vol5'])
temp6 = pd.DataFrame([0],columns=['vol_imbalance5'])
df_sub_book_feats5 = pd.concat([times_pd,temp,temp2,temp3,temp4,temp5,temp6],axis=1)
list_fin.append(df_sub_book_feats)
list_fin2.append(df_sub_book_feats5)
# Compute entropy
isEmpty = book_stock.query(f'seconds_in_bucket >= 480').empty
if isEmpty == False:
df_ent = book_stock.query(f'seconds_in_bucket >= 480').groupby(['time_id']).apply(entropy_from_df).to_frame().reset_index().fillna(0)
df_ent2 = book_stock.query(f'seconds_in_bucket >= 480').groupby(['time_id']).apply(entropy_from_df2).to_frame().reset_index().fillna(0)
df_ent3 = book_stock.query(f'seconds_in_bucket >= 480').groupby(['time_id']).apply(entropy_from_df3).to_frame().reset_index().fillna(0)
df_ent['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_ent['time_id']]
df_ent = df_ent.rename(columns={'time_id':'row_id',0:'entropy'})
df_ent2 = df_ent2.rename(columns={0:'entropy2'}).drop(['time_id'],axis=1)
df_ent3 = df_ent3.rename(columns={0:'entropy3'}).drop(['time_id'],axis=1)
df_ent = pd.concat([df_ent,df_ent2,df_ent3],axis=1)
else:
times_pd = pd.DataFrame(all_time_ids_byStock,columns=['time_id'])
times_pd['time_id'] = [f'{stock_id}-{time_id}' for time_id in times_pd['time_id']]
times_pd = times_pd.rename(columns={'time_id':'row_id'})
temp = pd.DataFrame([0],columns=['entropy'])
temp2 = pd.DataFrame([0],columns=['entropy2'])
temp3 = pd.DataFrame([0],columns=['entropy3'])
df_ent = pd.concat([times_pd,temp,temp2,temp3],axis=1)
list_ent.append(df_ent)
# Compute other metrics
df_others = book_stock.groupby(['time_id']).apply(other_metrics).to_frame().reset_index().fillna(0)
df_others = df_others.rename(columns={0:'embedding'})
df_others[['linearFit1_1','linearFit1_2','linearFit1_3','wap_std1_1','wap_std1_2','wap_std1_3']] = pd.DataFrame(df_others.embedding.tolist(), index=df_others.index)
df_others['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_others['time_id']]
df_others = df_others.rename(columns={'time_id':'row_id'}).drop(['embedding'],axis=1)
list_others.append(df_others)
isEmpty = book_stock.query(f'seconds_in_bucket >= 300').empty
if isEmpty == False:
df_others2 = book_stock.query(f'seconds_in_bucket >= 300').groupby(['time_id']).apply(other_metrics).to_frame().reset_index().fillna(0)
df_others2 = df_others2.rename(columns={0:'embedding'})
df_others2[['linearFit2_1','linearFit2_2','linearFit2_3','wap_std2_1','wap_std2_2','wap_std2_3']] = pd.DataFrame(df_others2.embedding.tolist(), index=df_others2.index)
df_others2['time_id'] = [f'{stock_id}-{time_id}' for time_id in df_others2['time_id']]
df_others2 = df_others2.rename(columns={'time_id':'row_id'}).drop(['embedding'],axis=1)
else:
times_pd = pd.DataFrame(all_time_ids_byStock,columns=['time_id'])
times_pd['time_id'] = [f'{stock_id}-{time_id}' for time_id in times_pd['time_id']]
times_pd = times_pd.rename(columns={'time_id':'row_id'})
temp = pd.DataFrame([0],columns=['linearFit2_1'])
temp2 = pd.DataFrame([0],columns=['linearFit2_2'])
temp3 = pd.DataFrame([0],columns=['linearFit2_3'])
temp4 = | pd.DataFrame([0],columns=['wap_std2_1']) | pandas.DataFrame |
import numpy as np
import pandas as pd
from ..mean_characters_per_word import MeanCharactersPerWord
from ..utils import PrimitiveT, find_applicable_primitives, valid_dfs
class TestMeanCharactersPerWord(PrimitiveT):
primitive = MeanCharactersPerWord
def test_sentences(self):
x = pd.Series(['This is a test file',
'This is second line',
'third line $1,000',
'and subsequent lines',
'and more'])
primitive_func = self.primitive().get_function()
answers = pd.Series([3.0, 4.0, 5.0, 6.0, 3.5])
pd.testing.assert_series_equal(primitive_func(x), answers, check_names=False)
def test_punctuation(self):
x = pd.Series(['This: is a test file',
'This, is second line?',
'third/line $1,000;',
'and--subsequen\'t lines...',
'*and, more..'])
primitive_func = self.primitive().get_function()
answers = | pd.Series([3.0, 4.0, 8.0, 10.5, 4.0]) | pandas.Series |
'''This module implements the word2vec model service that is responsible
for training the model as well as a backend interface for the API.
'''
from datetime import datetime
import json
import logging
import pandas as pd
from gensim.models.ldamulticore import LdaMulticore
import numpy as np
from wb_nlp.interfaces.milvus import (
get_milvus_client, get_embedding_dsl
)
from wb_nlp.interfaces import mongodb, elasticsearch
from wb_nlp.types.models import LDAModelConfig, ModelTypes
from wb_nlp.models.base import BaseModel
class LDAModel(BaseModel):
def __init__(
self,
model_config_id,
cleaning_config_id,
model_class=LdaMulticore,
model_config_type=LDAModelConfig,
expected_model_name=ModelTypes.lda.value,
model_run_info_description="",
model_run_info_id=None,
raise_empty_doc_status=True,
log_level=logging.INFO,
):
super().__init__(
model_config_id=model_config_id,
cleaning_config_id=cleaning_config_id,
model_class=model_class,
model_config_type=model_config_type,
expected_model_name=expected_model_name,
model_run_info_description=model_run_info_description,
model_run_info_id=model_run_info_id,
raise_empty_doc_status=raise_empty_doc_status,
log_level=log_level,
)
self.topic_composition_ranges = None
try:
self.logger.info("Initializing topic composition")
self.get_topic_composition_ranges()
self.logger.info("Finished initializing topic composition")
except ValueError:
self.logger.info("Skipping initialization of topic composition")
def set_model_specific_attributes(self):
self.num_topics = self.dim
def transform_doc(self, document, normalize=True, tolist=False):
# This function is the primary method of converting a document
# into its vector representation based on the model.
# This should be implemented in the respective models.
# The expected output is a dictionary with keys:
# - doc_vec # numpy array of shape (1, self.dim)
# - success # Whether the transformation went successfully
self.check_model()
success = True
document = document.lower()
try:
doc_topics = self.infer_topics(document)
doc_vec = np.array([dt["score"] for dt in sorted(
doc_topics, key=lambda x: x["topic"])]).reshape(1, -1)
if normalize:
doc_vec /= np.linalg.norm(doc_vec, ord=2)
except Exception as e:
success = False
if self.raise_empty_doc_status:
raise(e)
else:
doc_vec = np.zeros(self.dim).reshape(1, -1)
if tolist:
doc_vec = doc_vec.ravel().tolist()
return dict(doc_vec=doc_vec, success=success)
def infer_topics(self, text, topn_topics=None, total_topic_score=None, serialize=False):
if isinstance(text, str):
text = text.split()
if len(text) == 1:
doc_topics = self.model.get_term_topics(
self.g_dict.token2id[text[0]])
else:
doc = self.g_dict.doc2bow(text)
doc_topics = self.model[doc]
found_topics = {i for i, v in doc_topics}
# print(found_topics)
for i in range(self.model.num_topics):
if i not in found_topics:
doc_topics.append((i, 0))
doc_topics = pd.DataFrame(doc_topics, columns=['topic', 'score'])
doc_topics = doc_topics.sort_values('score', ascending=False)
if total_topic_score is not None:
tdoc_topics = doc_topics[doc_topics.score.cumsum(
) <= total_topic_score]
if tdoc_topics.empty:
doc_topics = doc_topics.head(1)
else:
doc_topics = tdoc_topics
if topn_topics is not None and doc_topics.shape[0] > topn_topics:
doc_topics = doc_topics.head(topn_topics)
# doc_topics['topic'] = doc_topics['topic'].astype(int)
doc_topics = doc_topics.to_dict('records')
if serialize:
doc_topics = json.dumps(doc_topics)
return doc_topics
def get_model_topic_words(self, topn_words=5, total_word_score=None, serialize=False):
payload = []
for topic_id in range(self.model.num_topics):
topic_words = self.get_topic_words(
topic_id,
topn_words=topn_words,
total_word_score=total_word_score
)
payload.append({'topic_id': topic_id, 'topic_words': topic_words})
if serialize:
payload = json.dumps(payload)
return payload
def get_topic_words(self, topic_id, topn_words=10, total_word_score=None, serialize=False):
topic_id = int(topic_id)
topic_words = pd.DataFrame(self.model.show_topic(
topic_id, topn=topn_words), columns=['word', 'score'])
topic_words = topic_words.sort_values('score', ascending=False)
if total_word_score is not None:
ttopic_words = topic_words[topic_words.score.cumsum(
) <= total_word_score]
if ttopic_words.empty:
topic_words = topic_words.head(1)
else:
topic_words = ttopic_words
if topn_words is not None and topic_words.shape[0] > topn_words:
topic_words = topic_words.head(topn_words)
topic_words = topic_words.to_dict('records')
if serialize:
topic_words = json.dumps(topic_words)
return topic_words
def get_doc_topic_words(self, text, topn_topics=10, topn_words=10, total_topic_score=1, total_word_score=1, serialize=False):
doc_topics = self.infer_topics(
text, topn_topics=topn_topics, total_topic_score=total_topic_score)
doc_topic_words = []
for dt in doc_topics:
topic = dt['topic']
topic_score = dt['score']
topic_words = self.get_topic_words(
topic, topn_words=topn_words, total_word_score=total_word_score)
topic_data = {'topic': topic, 'score': topic_score}
topic_data['words'] = topic_words
doc_topic_words.append(topic_data)
doc_topic_words = pd.DataFrame(doc_topic_words).to_dict('records')
if serialize:
doc_topic_words = json.dumps(doc_topic_words)
return doc_topic_words
# def get_doc_topic_words_by_id(self, doc_id, topn_topics=10, topn_words=10, total_topic_score=1, total_word_score=1, serialize=False):
# doc_topics = self.get_doc_topic_by_id(
# doc_id, topn=topn_topics, serialize=False)
# doc_topic_words = []
# for dt in doc_topics:
# topic = dt['topic']
# topic_score = dt['score']
# topic_words = self.get_topic_words(
# topic, topn_words=topn_words, total_word_score=total_word_score)
# topic_data = {'topic': topic, 'score': topic_score}
# topic_data['words'] = topic_words
# doc_topic_words.append(topic_data)
# doc_topic_words = pd.DataFrame(doc_topic_words).to_dict('records')
# if serialize:
# doc_topic_words = json.dumps(doc_topic_words)
# return doc_topic_words
def get_combined_doc_topic_words(self, text, topn_topics=None, topn_words=None, total_topic_score=0.8, total_word_score=0.2, serialize=False):
doc_topics = self.infer_topics(
text, topn_topics=topn_topics, total_topic_score=total_topic_score)
doc_topic_words = []
for dt in doc_topics:
topic = dt['topic']
topic_score = dt['score']
topic_words = self.get_topic_words(
topic, topn_words=topn_words, total_word_score=total_word_score)
for tw in topic_words:
word = tw['word']
word_score = tw['score']
doc_topic_words.append({
'topic': topic,
'word': word,
'topic_score': topic_score,
'word_score': word_score,
'score': topic_score * word_score
})
doc_topic_words = pd.DataFrame(doc_topic_words).sort_values(
'score', ascending=False).to_dict('records')
if serialize:
doc_topic_words = json.dumps(doc_topic_words)
return doc_topic_words
# def get_doc_topic_by_id(self, doc_id, topn=None, serialize=False):
# doc = self.documents_topics.loc[doc_id]
# if doc.empty:
# return []
# # Just call is score for consistency
# doc.name = 'score'
# doc.index.name = 'topic'
# doc = doc.sort_values(ascending=False) / doc.sum()
# doc.index = doc.index.astype(int)
# doc = doc.reset_index()
# if topn is not None:
# doc = doc.head(topn)
# doc = doc.to_dict('records')
# if serialize:
# doc = json.dumps(doc)
# return doc
# def get_similar_documents(self, document, topn=10, return_data='id', return_similarity=False, duplicate_threshold=0.01, show_duplicates=True, serialize=False):
# doc_topics = self.infer_topics(document)
# doc_topics = pd.DataFrame(doc_topics).sort_values(
# 'topic').set_index('topic')
# e_distance = euclidean_distances(doc_topics.score.values.reshape(
# 1, -1), self.normalized_documents_topics.values).flatten()
# if not show_duplicates:
# e_distance[e_distance <= duplicate_threshold] = np.inf
# payload = []
# for rank, top_sim_ix in enumerate(e_distance.argsort()[:topn], 1):
# payload.append(
# {'id': self.normalized_documents_topics.iloc[top_sim_ix].name, 'score': e_distance[top_sim_ix], 'rank': rank})
# if serialize:
# payload = pd.DataFrame(payload).to_json()
# return payload
# def get_similar_docs_by_id(self, doc_id, topn=10, return_data='id', return_similarity=False, duplicate_threshold=0.01, show_duplicates=True, serialize=False):
# doc_topics = self.normalized_documents_topics.loc[doc_id].values.reshape(
# 1, -1)
# e_distance = euclidean_distances(
# doc_topics, self.normalized_documents_topics.values).flatten()
# if not show_duplicates:
# e_distance[e_distance <= duplicate_threshold] = pd.np.inf
# payload = []
# for rank, top_sim_ix in enumerate(e_distance.argsort()[:topn], 1):
# payload.append(
# {'id': self.normalized_documents_topics.iloc[top_sim_ix].name, 'score': e_distance[top_sim_ix], 'rank': rank})
# if serialize:
# payload = pd.DataFrame(payload).to_json()
# return payload
def _mdb_get_docs_by_topic_composition(self, topic_percentage, return_all_topics=False):
self.check_wvecs()
model_run_info_id = self.model_run_info["model_run_info_id"]
doc_topic_collection = mongodb.get_document_topics_collection()
topic_cols = [f"topic_{id}" for id in sorted(topic_percentage)]
topic_filters = {f"topics.topic_{id}": {"$gte": val}
for id, val in sorted(topic_percentage.items())}
topic_filters = {
"model_run_info_id": model_run_info_id, **topic_filters}
cands = doc_topic_collection.find(topic_filters)
doc_df = pd.DataFrame(cands)
if doc_df.empty:
return doc_df
doc_df = doc_df.set_index(
"id")["topics"].apply(pd.Series)
if not return_all_topics:
doc_df = doc_df[topic_cols]
doc_df = doc_df.round(5)
return doc_df
def _get_docs_by_topic_composition_search(self, topic_percentage):
model_run_info_id = self.model_run_info["model_run_info_id"]
search = elasticsearch.DocTopic.search()
topic_filters = [dict(range={f"topics.topic_{id}": {"gte": val}})
for id, val in sorted(topic_percentage.items())]
search = search.query(
dict(
bool=dict(
must=[
{"term": {"model_run_info_id": model_run_info_id}}] + topic_filters
)
)
)
return search
def _get_docs_by_topic_composition(self, topic_percentage, return_all_topics=False):
self.check_wvecs()
search = self._get_docs_by_topic_composition_search(topic_percentage)
topic_cols = [f"topic_{id}" for id in sorted(topic_percentage)]
search = search[0: search.count()]
response = search.execute()
doc_df = pd.DataFrame([h.to_dict() for h in response.hits])
if doc_df.empty:
return doc_df
doc_df = doc_df.set_index(
"id")["topics"].apply(pd.Series)
if not return_all_topics:
doc_df = doc_df[topic_cols]
doc_df = doc_df.round(5)
return doc_df
def get_docs_by_topic_composition_count(self, topic_percentage):
search = self._get_docs_by_topic_composition_search(topic_percentage)
return search.count()
# return len(self._get_docs_by_topic_composition(topic_percentage))
def get_docs_by_topic_composition(self, topic_percentage, serialize=False, from_result=0, size=10, return_all_topics=False):
'''
topic_percentage (dict): key (int) corresponds to topic id and value (float [0, 1]) corresponds to the expected topic percentage.
'''
# topic_percentage = {6: 0.1, 42: 0.1}
doc_df = self._get_docs_by_topic_composition(
topic_percentage, return_all_topics=return_all_topics)
doc_count = len(doc_df)
payload = []
if doc_count > 0:
doc_rank = (-1 * doc_df
).rank().mean(axis=1).rank().sort_values()
doc_rank.name = "rank"
doc_rank = doc_rank.reset_index().to_dict("records")
doc_topic = doc_df.T.to_dict()
for rank, ent in enumerate(doc_rank):
if from_result > rank:
continue
payload.append(
{'id': ent["id"], 'topic': doc_topic[ent["id"]], 'rank': rank + 1})
if len(payload) == size:
break
payload = sorted(payload, key=lambda x: x['rank'])
if serialize:
payload = pd.DataFrame(payload).to_json()
return dict(total=doc_count, hits=payload)
def mdb_get_topic_composition_ranges(self, serialize=False):
self.check_wvecs()
if self.topic_composition_ranges is None:
model_run_info_id = self.model_run_info["model_run_info_id"]
doc_topic_collection = mongodb.get_document_topics_collection()
self.topic_composition_ranges = pd.DataFrame([
list(doc_topic_collection.aggregate([
{"$match": {"model_run_info_id": model_run_info_id}},
{"$group": {
"_id": f"topic_{i}",
"min": {"$min": f"$topics.topic_{i}"},
"max": {"$max": f"$topics.topic_{i}"}}}]))[0] for i in range(self.dim)
]).rename(columns={"_id": "topic"}).set_index("topic").T.to_dict() # "records")
# Data structure: {topic_id: {min: <min_val>, max: <max_val>}}
payload = self.topic_composition_ranges
if serialize:
payload = | pd.DataFrame(payload) | pandas.DataFrame |
import numpy as np
import pandas as pd
import pytest
from hypothesis import given, settings
from pandas.testing import assert_frame_equal
from janitor.testing_utils.strategies import (
conditional_df,
conditional_right,
conditional_series,
)
@pytest.mark.xfail(reason="empty object will pass thru")
@given(s=conditional_series())
def test_df_empty(s):
"""Raise ValueError if `df` is empty."""
df = pd.DataFrame([], dtype="int", columns=["A"])
with pytest.raises(ValueError):
df.conditional_join(s, ("A", "non", "=="))
@pytest.mark.xfail(reason="empty object will pass thru")
@given(df=conditional_df())
def test_right_empty(df):
"""Raise ValueError if `right` is empty."""
s = pd.Series([], dtype="int", name="A")
with pytest.raises(ValueError):
df.conditional_join(s, ("A", "non", "=="))
@given(df=conditional_df())
def test_right_df(df):
"""Raise TypeError if `right` is not a Series/DataFrame."""
with pytest.raises(TypeError):
df.conditional_join({"non": [2, 3, 4]}, ("A", "non", "=="))
@given(df=conditional_df(), s=conditional_series())
def test_right_series(df, s):
"""Raise ValueError if `right` is not a named Series."""
with pytest.raises(ValueError):
df.conditional_join(s, ("A", "non", "=="))
@given(df=conditional_df())
def test_df_MultiIndex(df):
"""Raise ValueError if `df` columns is a MultiIndex."""
with pytest.raises(ValueError):
df.columns = [list("ABCDE"), list("FGHIJ")]
df.conditional_join(
pd.Series([2, 3, 4], name="A"), (("A", "F"), "non", "==")
)
@given(df=conditional_df())
def test_right_MultiIndex(df):
"""Raise ValueError if `right` columns is a MultiIndex."""
with pytest.raises(ValueError):
right = df.copy()
right.columns = [list("ABCDE"), list("FGHIJ")]
df.conditional_join(right, (("A", "F"), "non", ">="))
@given(df=conditional_df(), s=conditional_series())
def test_check_conditions_exist(df, s):
"""Raise ValueError if no condition is provided."""
with pytest.raises(ValueError):
s.name = "B"
df.conditional_join(s)
@given(df=conditional_df(), s=conditional_series())
def test_check_condition_type(df, s):
"""Raise TypeError if any condition in conditions is not a tuple."""
with pytest.raises(TypeError):
s.name = "B"
df.conditional_join(s, ("A", "B", ""), ["A", "B"])
@given(df=conditional_df(), s=conditional_series())
def test_check_condition_length(df, s):
"""Raise ValueError if any condition is not length 3."""
with pytest.raises(ValueError):
s.name = "B"
df.conditional_join(s, ("A", "B", "C", "<"))
df.conditional_join(s, ("A", "B", ""), ("A", "B"))
@given(df=conditional_df(), s=conditional_series())
def test_check_left_on_type(df, s):
"""Raise TypeError if left_on is not a string."""
with pytest.raises(TypeError):
s.name = "B"
df.conditional_join(s, (1, "B", "<"))
@given(df=conditional_df(), s=conditional_series())
def test_check_right_on_type(df, s):
"""Raise TypeError if right_on is not a string."""
with pytest.raises(TypeError):
s.name = "B"
df.conditional_join(s, ("B", 1, "<"))
@given(df=conditional_df(), s=conditional_series())
def test_check_op_type(df, s):
"""Raise TypeError if the operator is not a string."""
with pytest.raises(TypeError):
s.name = "B"
df.conditional_join(s, ("B", "B", 1))
@given(df=conditional_df(), s=conditional_series())
def test_check_column_exists_df(df, s):
"""
Raise ValueError if `left_on`
can not be found in `df`.
"""
with pytest.raises(ValueError):
s.name = "B"
df.conditional_join(s, ("C", "B", "<"))
@given(df=conditional_df(), s=conditional_series())
def test_check_column_exists_right(df, s):
"""
Raise ValueError if `right_on`
can not be found in `right`.
"""
with pytest.raises(ValueError):
s.name = "B"
df.conditional_join(s, ("B", "A", ">="))
@given(df=conditional_df(), s=conditional_series())
def test_check_op_correct(df, s):
"""
Raise ValueError if `op` is not any of
`!=`, `<`, `>`, `>=`, `<=`.
"""
with pytest.raises(ValueError):
s.name = "B"
df.conditional_join(s, ("B", "B", "=!"))
@given(df=conditional_df(), s=conditional_series())
def test_check_how_type(df, s):
"""
Raise TypeError if `how` is not a string.
"""
with pytest.raises(TypeError):
s.name = "B"
df.conditional_join(s, ("B", "B", "<"), how=1)
@given(df=conditional_df(), s=conditional_series())
def test_check_how_value(df, s):
"""
Raise ValueError if `how` is not one of
`inner`, `left`, or `right`.
"""
with pytest.raises(ValueError):
s.name = "B"
df.conditional_join(s, ("B", "B", "<"), how="INNER")
@given(df=conditional_df(), right=conditional_right())
def test_dtype_strings_non_equi(df, right):
"""
Raise ValueError if the dtypes are both strings
on a non-equi operator.
"""
with pytest.raises(ValueError):
df.conditional_join(right, ("C", "Strings", "<"))
@given(df=conditional_df(), s=conditional_series())
def test_dtype_not_permitted(df, s):
"""
Raise ValueError if dtype of column in `df`
is not an acceptable type.
"""
df["F"] = pd.Timedelta("1 days")
with pytest.raises(ValueError):
s.name = "A"
df.conditional_join(s, ("F", "A", "<"))
@given(df=conditional_df(), s=conditional_series())
def test_dtype_str(df, s):
"""
Raise ValueError if dtype of column in `df`
does not match the dtype of column from `right`.
"""
with pytest.raises(ValueError):
s.name = "A"
df.conditional_join(s, ("C", "A", "<"))
@given(df=conditional_df(), s=conditional_series())
def test_dtype_category_non_equi(df, s):
"""
Raise ValueError if dtype is category,
and op is non-equi.
"""
with pytest.raises(ValueError):
s.name = "A"
s = s.astype("category")
df["C"] = df["C"].astype("category")
df.conditional_join(s, ("C", "A", "<"))
@given(df=conditional_df(), s=conditional_series())
def test_check_sort_by_appearance_type(df, s):
"""
Raise TypeError if `sort_by_appearance` is not a boolean.
"""
with pytest.raises(TypeError):
s.name = "B"
df.conditional_join(s, ("B", "B", "<"), sort_by_appearance="True")
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_less_than_floats(df, right):
"""Test output for a single condition. "<"."""
left_on, right_on = ["B", "Numeric"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} < {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "<"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_less_than_ints(df, right):
"""Test output for a single condition. "<"."""
left_on, right_on = ["A", "Integers"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1, C="2"), on="t")
.query(f"{left_on} < {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "<"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_less_than_ints_extension_array(df, right):
"""Test output for a single condition. "<"."""
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
left_on, right_on = ["A", "Integers"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} < {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "<"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_less_than_equal(df, right):
"""Test output for a single condition. "<=". DateTimes"""
left_on, right_on = ["E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} <= {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "<="), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_less_than_date(df, right):
"""Test output for a single condition. "<". Dates"""
left_on, right_on = ["E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} < {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "<"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_greater_than_datetime(df, right):
"""Test output for a single condition. ">". Datetimes"""
left_on, right_on = ["E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} > {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, ">"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_greater_than_ints(df, right):
"""Test output for a single condition. ">="."""
left_on, right_on = ["A", "Integers"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} >= {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, ">="), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_greater_than_floats_floats(df, right):
"""Test output for a single condition. ">"."""
left_on, right_on = ["B", "Numeric"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} > {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, ">"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_greater_than_ints_extension_array(df, right):
"""Test output for a single condition. ">="."""
left_on, right_on = ["A", "Integers"]
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} > {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, ">"), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_not_equal_numeric(df, right):
"""Test output for a single condition. "!="."""
left_on, right_on = ["A", "Integers"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.dropna(subset=["A", "Integers"])
.query(f"{left_on} != {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "!="), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_not_equal_ints_only(df, right):
"""Test output for a single condition. "!="."""
left_on, right_on = ["A", "Integers"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.dropna(subset=["A", "Integers"])
.query(f"{left_on} != {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "!="), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_not_equal_floats_only(df, right):
"""Test output for a single condition. "!="."""
left_on, right_on = ["B", "Numeric"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.dropna(subset=["B", "Numeric"])
.query(f"{left_on} != {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "!="), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_not_equal_datetime(df, right):
"""Test output for a single condition. "!="."""
left_on, right_on = ["E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.dropna(subset=["E", "Dates"])
.query(f"{left_on} != {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "!="), how="inner", sort_by_appearance=True
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_equality_string(df, right):
"""Test output for a single condition. "=="."""
left_on, right_on = ["C", "Strings"]
expected = df.dropna(subset=[left_on]).merge(
right.dropna(subset=[right_on]), left_on=left_on, right_on=right_on
)
expected = expected.reset_index(drop=True)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "=="), how="inner", sort_by_appearance=False
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@pytest.mark.xfail(
reason="""sometimes, categories are coerced to objects;
might be a pandas version issue.
"""
)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_equality_category(df, right):
"""Test output for a single condition. "=="."""
left_on, right_on = ["C", "Strings"]
df = df.assign(C=df["C"].astype("category"))
right = right.assign(Strings=right["Strings"].astype("category"))
expected = df.dropna(subset=[left_on]).merge(
right.dropna(subset=[right_on]), left_on=left_on, right_on=right_on
)
expected = expected.reset_index(drop=True)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "=="), how="inner", sort_by_appearance=False
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_equality_numeric(df, right):
"""Test output for a single condition. "=="."""
left_on, right_on = ["A", "Integers"]
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
df.loc[0, "A"] = pd.NA
right.loc[0, "Integers"] = pd.NA
expected = df.dropna(subset=[left_on]).merge(
right.dropna(subset=[right_on]), left_on=left_on, right_on=right_on
)
expected = expected.reset_index(drop=True)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "=="), how="inner", sort_by_appearance=False
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_single_condition_equality_datetime(df, right):
"""Test output for a single condition. "=="."""
left_on, right_on = ["E", "Dates"]
expected = df.dropna(subset=[left_on]).merge(
right.dropna(subset=[right_on]), left_on=left_on, right_on=right_on
)
expected = expected.reset_index(drop=True)
expected = expected.filter([left_on, right_on])
actual = df.conditional_join(
right, (left_on, right_on, "=="), how="inner", sort_by_appearance=False
)
actual = actual.filter([left_on, right_on])
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_how_left(df, right):
"""Test output when `how==left`. "<="."""
left_on, right_on = ["A", "Integers"]
expected = (
df.assign(t=1, index=np.arange(len(df)))
.merge(right.assign(t=1), on="t")
.query(f"{left_on} <= {right_on}")
)
expected = expected.set_index("index")
expected.index.name = None
expected = df.join(
expected.filter(right.columns), how="left", sort=False
).reset_index(drop=True)
actual = df.conditional_join(
right, (left_on, right_on, "<="), how="left", sort_by_appearance=True
)
assert_frame_equal(expected, actual)
@given(df=conditional_df(), right=conditional_right())
def test_how_right(df, right):
"""Test output when `how==right`. ">"."""
left_on, right_on = ["E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1, index=np.arange(len(right))), on="t")
.query(f"{left_on} > {right_on}")
)
expected = expected.set_index("index")
expected.index.name = None
expected = (
expected.filter(df.columns)
.join(right, how="right", sort=False)
.reset_index(drop=True)
)
actual = df.conditional_join(
right, (left_on, right_on, ">"), how="right", sort_by_appearance=True
)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_gt_and_lt_dates(df, right):
"""Test output for interval conditions."""
middle, left_on, right_on = ("E", "Dates", "Dates_Right")
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} < {middle} < {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, middle, right_on])
actual = df.conditional_join(
right,
(middle, left_on, ">"),
(middle, right_on, "<"),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([left_on, middle, right_on])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_ge_and_le_dates(df, right):
"""Test output for interval conditions."""
middle, left_on, right_on = ("E", "Dates", "Dates_Right")
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} <= {middle} <= {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, middle, right_on])
actual = df.conditional_join(
right,
(middle, left_on, ">="),
(middle, right_on, "<="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([left_on, middle, right_on])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_le_and_ge_dates(df, right):
"""Test output for interval conditions."""
middle, left_on, right_on = ("E", "Dates", "Dates_Right")
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} <= {middle} <= {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, middle, right_on])
actual = df.conditional_join(
right,
(middle, right_on, "<="),
(middle, left_on, ">="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([left_on, middle, right_on])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_ge_and_le_numbers(df, right):
"""Test output for interval conditions."""
middle, left_on, right_on = ("B", "Numeric", "Floats")
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} <= {middle} <= {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, middle, right_on])
actual = df.conditional_join(
right,
(middle, left_on, ">="),
(middle, right_on, "<="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([left_on, middle, right_on])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_le_and_ge_numbers(df, right):
"""Test output for interval conditions."""
middle, left_on, right_on = ("B", "Numeric", "Floats")
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} <= {middle} <= {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, middle, right_on])
actual = df.conditional_join(
right,
(middle, right_on, "<="),
(middle, left_on, ">="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([left_on, middle, right_on])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_gt_and_lt_numbers(df, right):
"""Test output for interval conditions."""
middle, left_on, right_on = ("B", "Numeric", "Floats")
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(f"{left_on} < {middle} < {right_on}")
.reset_index(drop=True)
)
expected = expected.filter([left_on, middle, right_on])
actual = df.conditional_join(
right,
(middle, left_on, ">"),
(middle, right_on, "<"),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([left_on, middle, right_on])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_gt_and_lt_numbers_(df, right):
"""
Test output for multiple conditions.
"""
first, second, third = ("Numeric", "Floats", "B")
expected = (
right.assign(t=1)
.merge(df.assign(t=1), on="t")
.query(f"{first} > {third} and {second} < {third}")
.reset_index(drop=True)
)
expected = expected.filter([first, second, third])
actual = right.conditional_join(
df,
(first, third, ">"),
(second, third, "<"),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter([first, second, third])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_gt_and_lt_numbers_left_join(df, right):
"""
Test output for multiple conditions, and how is `left`.
"""
first, second, third = ("Numeric", "Floats", "B")
right = right.assign(t=1, check=range(len(right)))
df = df.assign(t=1)
expected = right.merge(df, on="t").query(
f"{first} > {third} and {second} < {third}"
)
drop = right.columns.difference(["check"])
expected = right.merge(
expected.drop(columns=[*drop]), on="check", how="left", sort=False
)
expected = expected.filter([first, second, third])
actual = right.conditional_join(
df,
(first, third, ">"),
(second, third, "<"),
how="left",
sort_by_appearance=True,
)
actual = actual.droplevel(0, 1)
actual = actual.loc[:, [first, second, third]]
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_gt_and_lt_numbers_right_join(df, right):
"""
Test output for multiple conditions, and how is `right`.
"""
first, second, third = ("Numeric", "Floats", "B")
df = df.assign(t=1, check=range(len(df)))
right = right.assign(t=1)
expected = right.merge(df, on="t").query(
f"{first} > {third} and {second} < {third}"
)
drop = df.columns.difference(["check"])
expected = expected.drop(columns=[*drop]).merge(
df, on="check", how="right", sort=False
)
expected = expected.filter([first, second, third])
actual = right.conditional_join(
df,
(first, third, ">"),
(second, third, "<"),
how="right",
sort_by_appearance=True,
)
actual = actual.droplevel(0, 1)
actual = actual.loc[:, [first, second, third]]
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_ne(df, right):
"""
Test output for multiple conditions. `!=`
"""
filters = ["A", "Integers", "B", "Numeric"]
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A != Integers and B != Numeric")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("A", "Integers", "!="),
("B", "Numeric", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_ne_extension(df, right):
"""
Test output for multiple conditions. `!=`
"""
filters = ["A", "Integers", "B", "Numeric"]
df = df.assign(A=df["A"].astype("Int64"))
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A != Integers and B != Numeric")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("A", "Integers", "!="),
("B", "Numeric", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_ne_extension_right(df, right):
"""
Test output for multiple conditions. `!=`
"""
filters = ["A", "Integers", "B", "Numeric"]
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A != Integers and B != Numeric")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("A", "Integers", "!="),
("B", "Numeric", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_ne_dates(df, right):
"""
Test output for multiple conditions. `!=`
"""
filters = ["A", "Integers", "E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A != Integers and E != Dates")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("A", "Integers", "!="),
("E", "Dates", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_multiple_ne_dates(df, right):
"""
Test output for multiple conditions. `!=`
"""
filters = ["A", "Integers", "E", "Dates", "B", "Numeric"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A != Integers and E != Dates and B != Numeric")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("A", "Integers", "!="),
("E", "Dates", "!="),
("B", "Numeric", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_eq_and_ne(df, right):
"""Test output for equal and not equal conditions."""
A, B, C, D = ("B", "Numeric", "E", "Dates")
expected = (
df.merge(right, left_on=A, right_on=B)
.dropna(subset=[A, B])
.query(f"{C} != {D}")
.reset_index(drop=True)
)
expected = expected.filter([A, B, C, D])
actual = df.conditional_join(
right,
(A, B, "=="),
(C, D, "!="),
how="inner",
sort_by_appearance=False,
)
actual = actual.filter([A, B, C, D])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_conditions_ne_and_eq(df, right):
"""Test output for equal and not equal conditions."""
A, B, C, D = ("A", "Integers", "E", "Dates")
expected = (
df.merge(right, left_on=C, right_on=D)
.dropna(subset=[C, D])
.query(f"{A} != {B}")
.reset_index(drop=True)
)
expected = expected.filter([A, B, C, D])
actual = df.conditional_join(
right,
(A, B, "!="),
(C, D, "=="),
how="inner",
sort_by_appearance=False,
)
actual = actual.filter([A, B, C, D])
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_gt_lt_ne_conditions(df, right):
"""
Test output for multiple conditions.
"""
filters = ["A", "Integers", "B", "Numeric", "E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A > Integers and B < Numeric and E != Dates")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("A", "Integers", ">"),
("B", "Numeric", "<"),
("E", "Dates", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_gt_lt_ne_start(df, right):
"""
Test output for multiple conditions.
"""
filters = ["A", "Integers", "B", "Numeric", "E", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1, C="C"), on="t")
.query("A > Integers and B < Numeric and E != Dates")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("E", "Dates", "!="),
("A", "Integers", ">"),
("B", "Numeric", "<"),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_ge_le_ne_extension_array(df, right):
"""
Test output for multiple conditions.
"""
filters = ["A", "Integers", "B", "Numeric", "E", "Dates"]
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query("A != Integers and B < Numeric and E >= Dates")
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("E", "Dates", ">="),
("A", "Integers", "!="),
("B", "Numeric", "<"),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_ge_lt_ne_extension(df, right):
"""
Test output for multiple conditions.
"""
filters = ["A", "Integers", "B", "Numeric", "E", "Dates"]
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype(pd.Int64Dtype()))
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t")
.query(
"A < Integers and B != Numeric and E >= Dates and E != Dates_Right"
)
.reset_index(drop=True)
)
expected = expected.filter(filters)
actual = df.conditional_join(
right,
("E", "Dates", ">="),
("B", "Numeric", "!="),
("A", "Integers", "<"),
("E", "Dates_Right", "!="),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(filters)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_eq_ge_and_le_numbers(df, right):
"""Test output for multiple conditions."""
l_eq, l_ge, l_le = ["B", "A", "E"]
r_eq, r_ge, r_le = ["Floats", "Integers", "Dates"]
columns = ["B", "A", "E", "Floats", "Integers", "Dates"]
expected = (
df.merge(right, left_on=l_eq, right_on=r_eq, how="inner", sort=False)
.dropna(subset=[l_eq, r_eq])
.query(f"{l_ge} >= {r_ge} and {l_le} <= {r_le}")
.reset_index(drop=True)
)
expected = expected.filter(columns)
actual = df.conditional_join(
right,
(l_eq, r_eq, "=="),
(l_ge, r_ge, ">="),
(l_le, r_le, "<="),
how="inner",
sort_by_appearance=False,
)
actual = actual.filter(columns)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_dual_ge_and_le_diff_numbers(df, right):
"""Test output for multiple conditions."""
l_ge, l_le = ["A", "E"]
r_ge, r_le = ["Integers", "Dates"]
columns = ["B", "A", "E", "Floats", "Integers", "Dates"]
expected = (
df.assign(t=1)
.merge(right.assign(t=1), on="t", how="inner", sort=False)
.query(f"{l_ge} > {r_ge} and {l_le} <= {r_le}")
.reset_index(drop=True)
)
expected = expected.filter(columns)
actual = df.conditional_join(
right,
(l_le, r_le, "<="),
(l_ge, r_ge, ">"),
how="inner",
sort_by_appearance=True,
)
actual = actual.filter(columns)
assert_frame_equal(expected, actual)
@settings(deadline=None)
@given(df=conditional_df(), right=conditional_right())
def test_ge_lt_ne_extension_variant(df, right):
"""
Test output for multiple conditions.
"""
filters = ["A", "Integers", "B", "Numeric", "E", "Dates"]
df = df.assign(A=df["A"].astype("Int64"))
right = right.assign(Integers=right["Integers"].astype( | pd.Int64Dtype() | pandas.Int64Dtype |
# -*- coding: utf-8 -*-
"""
Created on Thu Jun 14 12:04:33 2018
@author: gurunath.lv
"""
try :
import base64
import datetime
import io
import dash
from dash.dependencies import Input, Output
import dash_core_components as dcc
import dash_html_components as html
import dash_table_experiments as dt
import plotly.graph_objs as go
import numpy as np
import pandas as pd
import json
# from tf_universal_sent_emb import get_similar_records
# from spacy_text_classifier_cnn import train_cnn_for_given_label,predict
import glob
import os
# from custom_classifier import customKNN,ParagraphVectors
# from dashboard import Dashboard
from flask import Flask
import flask
#import glob
from sklearn.pipeline import Pipeline
import pickle
from lime.lime_text import LimeTextExplainer
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.feature_extraction.text import TfidfVectorizer
except ImportError as e:
print("some packages are not installed -to use this textclf Annotator \
! please install relevant libraries",e)
server = Flask(__name__)
app=dash.Dash(name = __name__, server = server)
if not os.path.exists('tmp'):
os.mkdir('tmp')
DIRECTORY_PATH=r'tmp\\'
#app = dash.Dash()
app.scripts.config.serve_locally = True
app.config['suppress_callback_exceptions']=True
# custom_dush=Dashboard()
#prodapt=html.Div(html.Img(src='http://www.prodapt.com/wp-content/uploads/logo_prodapt.png')),
#vs=html.H1('vs')
#reinfer=html.Div(html.Img(src='https://d1qb2nb5cznatu.cloudfront.net/startups/i/703763-82fa920eed7d56e7cdcee1b1d9a30b14-medium_jpg.jpg?buster=1440002957')),
#logo=custom_dush.three_columns_grid(prodapt,vs,reinfer)
def transform_using_tfidf(text_series):
tfidf=TfidfVectorizer(stop_words='english')
array=tfidf.fit_transform(text_series.tolist()).toarray()
return array,tfidf
def similarity_measure(inp_sent,array,tfidf,top_n):
inp_vec=tfidf.transform([inp_sent]).toarray()
cs=cosine_similarity(inp_vec,array)
top_match_index=np.flip(np.argsort(cs,axis=1)[:,-top_n:],axis=1)
return top_match_index
def get_similar_records(inp_sent,total_text,top_n=10):
array,tfidf=transform_using_tfidf(total_text)
top_match_index=similarity_measure(inp_sent,array,tfidf,top_n)
return total_text.iloc[top_match_index.ravel()]
app.layout = html.Div([
# logo,
dcc.Upload(
id='upload-data',
children=html.Div([
'Drag and Drop or ',
html.A('Select Files')
]),
style={
'width': '100%',
'height': '60px',
'lineHeight': '60px',
'borderWidth': '1px',
'borderStyle': 'dashed',
'borderRadius': '5px',
'textAlign': 'center',
'margin': '10px'
},
# Allow multiple files to be uploaded
multiple=True
),
html.Div(id='output-data-upload'),
html.Div(dt.DataTable(rows=[{}]), style={'display': 'none'}),
dcc.Input(id='user-input-for-similarity',
value='Enter the sentence', type='text',
style={'width': '49%','align':'center'}),
html.Div(id='similar-docs'),
html.Br(),
html.H3('Training dataset'),
html.Div(id='output'),
html.Button('Train',id='train-button'),
html.Br(),
dcc.Input(id='user-input-for-prediction',
value='Enter the sentence to predict', type='text',
style={'width': '49%','align':'center'}),
html.H1(id='train-output'),
# html.Button('Del data',id='delete-button'),
html.H1(id='del-output'),
dcc.Graph(id='predict-output'),
html.Br(),
dcc.Link('Why ML made this Prediction !!!!', href='/explain'),
# html.Div([
# html.Pre(id='output', className='two columns'),
# html.Div(
# dcc.Graph(
# id='graph',
# style={
# 'overflow-x': 'wordwrap'
# }
# ),
# className='ten columns'
# )
# ], className='row')
])
def parse_contents(contents, file_name, date):
content_type, content_string = contents.split(',')
decoded = base64.b64decode(content_string)
global df
global filename
try:
if 'csv' in file_name:
# Assume that the user uploaded a CSV file
df = pd.read_csv(
io.StringIO(decoded.decode('utf8')))
elif 'xls' in file_name:
# Assume that the user uploaded an excel file
df = pd.read_excel(io.BytesIO(decoded))
except Exception as e:
print(e)
return html.Div([
'There was an error processing this file.'
])
"""
pass similar contents df not file uploaded df
To be filled
"""
# df.to_csv(r'{}'.format(filename),index=False)
filename=file_name
return html.Div([
html.H5(filename),
html.H6(datetime.datetime.fromtimestamp(date)),
# Use the DataTable prototype component:
# github.com/plotly/dash-table-experiments
dt.DataTable(rows=df.to_dict('records'),id='edit-table'),
html.Hr(), # horizontal line
# For debugging, display the raw contents provided by the web browser
# html.Div('Raw Content'),
# html.Pre(contents[0:200] + '...', style={
# 'whiteSpace': 'pre-wrap',
# 'wordBreak': 'break-all'
# })
])
@app.callback(
Output(component_id='similar-docs', component_property='children'),
[Input(component_id='user-input-for-similarity', component_property='value')])
def get_similar_docs(sent):
print('similar docs called ',sent)
# path=glob.glob(r'*.csv')
# df=pd.read_csv(path[0],encoding='ISO-8859-1')
global similar_df
similar_series=get_similar_records(sent,df[df.columns[0]])
similar_df=pd.DataFrame(columns=['Similar_sentences','labels'])
similar_df['Similar_sentences']=similar_series
print('check',similar_df.head())
# similar_df.to_csv
return html.Div(dt.DataTable(rows=similar_df.to_dict('records'),id='edit-table-similar'),)
def train_custom_classifier(similar_df,filename):
texts, labels =similar_df.iloc[:,0].values,similar_df.iloc[:,1].values
print(type(labels),type(texts),labels)
if glob.glob(r'{}{}_knn_centroid.pkl'.format(DIRECTORY_PATH,filename)):
dict_=pickle.load(open(glob.glob(r'{}*{}_knn_centroid.pkl'.format(DIRECTORY_PATH,filename))[0],'rb'))
pipe=Pipeline(steps=[('pv',ParagraphVectors(filename=filename)),('knn',customKNN(label_to_vect_dict=dict_))])
label_encoding=pickle.load(open(glob.glob(r'{}*{}_label_encoding.pkl'.format(DIRECTORY_PATH,filename))[0],'rb'))
for idx,lab in enumerate(set(labels)):
label_encoding[idx+len(label_encoding)]=lab
else:
label_encoding=dict()
pipe=Pipeline(steps=[('pv',ParagraphVectors(filename=filename)),('knn',customKNN())])
for idx,lab in enumerate(set(labels)):
label_encoding[idx]=lab
look_up=dict()
for k,v in label_encoding.items():
look_up[v]=k
pipe.fit(texts, | pd.Series(labels) | pandas.Series |
# -*- coding: utf-8 -*-
""" Calculate isotopic interference and standard ratios. """
import pandas as pd
import itertools
from interference_calculator.molecule import Molecule, mass_electron, periodic_table
"""xmin = 0.0
xmax = 0.0"""
def interference(atoms, target, targetrange=0.3, maxsize=5, charge=[1],
chargesign='-', style='plain'):
""" For a list of atoms (the composition of the sample),
calculate all molecules that can be formed from a
combination of those atoms (the interferences),
including all stable isotopes, up to maxsize atoms,
that have a mass-to-charge ratio within target ± targetrange.
The target can be given as a mass-to-charge ratio or as a
molecular formula. Molecular formulas are interpreted by Molecule().
See Molecule() docstring for a detailed explanation on how to enter
molecular formulas. If target is None, no filtering will be done and
all possible combinations of all atoms and isotopes up to maxsize
length will be calculated. Target information will be added to the
output, unless target is None.
Charge is usually 1, irrespective of sign. Give charge = [1, 2, 3]
to also include interferences with higher charges. Masses are
adjusted for missing electrons (+ charge), extra electrons (- charge),
or not adjusted (o charge, lower-case letter O). Setting charge=0
has the same effect as setting chargesign='o'. The charge for the
target ion, if target is specified as molecule instead of a number,
can be different from the charge on the interferences. If no charge is
specified for the target, the first charge and the chargesign of the
interferences are used for the target.
Molecular formulas are formatted in style (default is 'plain').
See Molecule() for more options.
Returns a pandas.DataFrame with a column 'molecule' with molecular formula,
a column 'charge', a column 'mass/charge' for the mass-to-charge ratio, a
column 'mass/charge diff' for the mass/charge difference between this ion
and the target mass/charge, a column 'MRP' which gives the mass-resolving
power (mz/Δmz) needed to resolve this ion from the target ion, a column
'target', which indicates whether this row was specified as the target,
and a column 'probability', which gives the combinatorial probability of
encoutering this combination of isotopes, given the composition of the
sample and the natural abundances of the isotopes.
"""
if isinstance(charge, (int, float, str)):
charge = tuple(int(charge))
elif isinstance(charge, (tuple, list)):
charge = tuple(int(c) for c in charge)
else:
raise ValueError('charge must be given as a number or a list of numbers.')
if chargesign not in ('+', '-', 'o', '0'):
raise ValueError('chargesign must be either "+", "-", "o", or "0".')
# How to handle charge?
# 1. charge for interferences
# - can be multiple values
# - specified by parameter
# 2. charge for target
# - only one value
# - can be different from 1
# - must be specified in target formula
# - if unspecified, take sign and first value from 1
if target:
try:
target_mz = float(target)
target = str(target)
target_charge = 0
target_chargesign = 'o'
target_abun = 1
except ValueError:
m = Molecule(target)
inferred_charge = False
if m.chargesign:
target_chargesign = m.chargesign
else:
target_chargesign = chargesign
inferred_charge = True
if m.charge:
target_charge = m.charge
else:
target_charge = charge[0]
inferred_charge = True
# If no charge was specified on target,
# push the inferred charge back to target
if inferred_charge:
if target_charge == 0:
pass
elif target_charge == 1:
target += ' {}'.format(target_chargesign)
else:
target += ' {}{}'.format(target_charge, target_chargesign)
target_mz = m.mass
target_abun = m.abundance
if m.charge > 0:
# mass correction done in Molecule.parse()
target_mz /= m.charge
else:
target_mz = 0
target_charge = 0
target_chargesign = '0'
target_abun = 0
# Retrieve info from perioic table for all atoms in sample.
# Create a list with all possible combinations up to maxsize atoms.
# Create same list for masses, combos are created in same order.
picked_atoms = periodic_table[periodic_table['element'].isin(atoms)]
isotope_combos = []
mass_combos = []
for size in range(1, maxsize + 1):
i = itertools.combinations_with_replacement(picked_atoms['isotope'], size)
m = itertools.combinations_with_replacement(picked_atoms['mass'], size)
isotope_combos.extend(list(i))
mass_combos.extend(list(m))
masses = pd.DataFrame(mass_combos).sum(axis=1)
molecules = [' '.join(m) for m in isotope_combos]
data = pd.DataFrame({'molecule': molecules,
'mass/charge': masses})
# ignore charge(s) for sign o
if chargesign in ('o', '0'):
data['charge'] = 0
else:
data_w_charge = []
for ch in charge:
d = data.copy()
d['charge'] = ch
if ch == 0:
data_w_charge.append(d)
continue
elif ch == 1:
charge_str = ' {}'.format(chargesign)
else:
charge_str = ' {}{}'.format(ch, chargesign)
d['molecule'] += charge_str
d['mass/charge'] /= ch
if chargesign == '+':
d['mass/charge'] -= mass_electron
else:
d['mass/charge'] += mass_electron
data_w_charge.append(d)
data = | pd.concat(data_w_charge) | pandas.concat |
import string
from typing import Any, Dict, List, Optional, Tuple, Mapping, Callable, Union
import pandas as pd
import numpy as np
import pytest
def _resolve_random_state(random_state: Union[int, np.random.RandomState]) -> np.random.RandomState:
""" Return a RandomState based on Input Integer (as seed) or RandomState"""
if isinstance(random_state, int):
return np.random.RandomState(random_state)
elif isinstance(random_state, np.random.RandomState):
return random_state
else:
raise NotImplementedError(
f"The random_state must be an integer or np.random.RandomState, "
f"current type: {type(random_state)}"
)
def _gen_random_int_series(
size: int, low: int = -100, high: int = 100, random_state: Union[int, np.random.RandomState] = 0
) -> pd.Series:
"""Return a randonly generated int Series, where the value is in [low, high]"""
rand = _resolve_random_state(random_state)
arr = rand.randint(low=low, high=high, size=size)
return pd.Series(arr)
def _gen_random_float_series(
size: int, random_state: Union[int, np.random.RandomState] = 0
) -> pd.Series:
"""Return a randonly generated float Series, with normal distribution"""
rand = _resolve_random_state(random_state)
arr = rand.normal(size=size)
return pd.Series(arr)
def _gen_random_bool_series(
size: int, random_state: Union[int, np.random.RandomState] = 0
) -> pd.Series:
"""Return a randonly generated boolean Series"""
rand = _resolve_random_state(random_state)
arr = rand.choice([True, False], size=size)
return pd.Series(arr)
def _gen_random_datatime_series(
size: int,
start: str = "1/1/2018",
end: str = "1/1/2019",
random_state: Union[int, np.random.RandomState] = 0,
) -> pd.Series:
"""Return a randonly generated datetime Series, where time in [start, end]"""
rand = _resolve_random_state(random_state)
population = pd.date_range(start, end)
arr = rand.choice(population, size=size)
return pd.Series(arr)
def _gen_random_string_series(
size: int,
min_len: int = 1,
max_len: int = 100,
random_state: Union[int, np.random.RandomState] = 0,
) -> pd.Series:
"""Return a randonly generated string Series, where string length is in [min_len, max_len]"""
rand = _resolve_random_state(random_state)
population = list(string.printable)
lst = []
for _ in range(size):
curr_len = rand.randint(min_len, max_len)
randstr = "".join(rand.choice(population, size=curr_len))
lst.append(randstr)
return pd.Series(lst)
def gen_constant_series(size: int, value: Any) -> pd.Series:
"""Return a constant pd.Series with given size and fill in given value"""
return pd.Series(value, index=range(size))
def gen_random_series(
size: int,
dtype: str = "object",
na_ratio: float = 0.0,
str_max_len: int = 100,
random_state: Union[int, np.random.RandomState] = 0,
) -> pd.Series:
"""
Return a randomly generated Pandas Series.
Parameters
----------
size: int
The size of the generated series
dtype: string
The type of the generated series.
Chosen from 'int', 'float', 'boolean', 'datetime', 'string' and 'object'.
na_ratio: float
The ratio of NA values in the series. Should be in [0.0, 1.0]
str_max_len: int
The max len of random string
seed: int
generator seed
"""
gen_func: Mapping[str, Callable[..., pd.Series]] = {
"int": _gen_random_int_series,
"float": _gen_random_float_series,
"boolean": _gen_random_bool_series,
"datetime": _gen_random_datatime_series,
"string": _gen_random_string_series,
}
if (dtype not in gen_func) and dtype != "object":
raise NotImplementedError(f"dtype {dtype} generator is not implemented.")
rand = _resolve_random_state(random_state)
# Generate non-NA series then replace some with NA.
# This can keep the type as the original type rather than object.
population_list = []
for curr_type in gen_func:
if dtype in [curr_type, "object"]:
if curr_type != "string":
rand_series = gen_func[curr_type](size, random_state=rand)
else:
rand_series = gen_func[curr_type](size, max_len=str_max_len, random_state=rand)
population_list.append(rand_series)
object_population = pd.concat(population_list, ignore_index=True)
object_series = pd.Series(rand.choice(object_population, size=size))
# Replace some values with NA.
na_pos = object_series.sample(frac=na_ratio, random_state=rand).index
if not na_pos.empty:
object_series[na_pos] = np.nan
return object_series
def gen_random_dataframe(
nrows: int = 30,
ncols: int = 30,
na_ratio: float = 0.0,
str_col_name_max_len: int = 100,
random_state: Union[int, np.random.RandomState] = 0,
) -> pd.DataFrame:
"""
Return a randomly generated dataframe.
The column name, data types are both randomly generated.
Note that if na_ratio is not 0.0, then the column type may not contain all types,
since there is a type transform when add NA to some series, e.g., boolean.
Parameters
----------
nrows: int
Number of rows of the generated dataframe.
na_ratio:
Ratio of NA values.
str_col_name_max_len:
max length of string column name
ncols: int
Number of columns of the generated dataframe.
seed: int
Random Seed
"""
rand = _resolve_random_state(random_state)
dtypes = ["int", "float", "boolean", "datetime", "string", "object"]
# Generate random columns
col_types = rand.choice(dtypes, size=ncols)
series_list = {}
for i in range(ncols):
series = gen_random_series(nrows, dtype=col_types[i], na_ratio=na_ratio, random_state=rand)
series_list[i] = series
df = pd.DataFrame(series_list)
# Generate random column names and index.
col_names = gen_random_series(
size=ncols,
dtype="object",
na_ratio=0.1,
str_max_len=str_col_name_max_len,
random_state=rand,
)
df.columns = col_names
df.index = gen_random_series(
df.index.shape[0], na_ratio=0.1, str_max_len=str_col_name_max_len, random_state=rand
)
return df
def gen_test_df() -> pd.DataFrame:
rand = np.random.RandomState(0)
nrows = 30
data = {}
data[0] = gen_random_dataframe(nrows=nrows, ncols=10, random_state=rand).reset_index(drop=True)
data[1] = gen_random_dataframe(
nrows=nrows, ncols=10, na_ratio=0.1, random_state=rand
).reset_index(drop=True)
data[2] = pd.Series([np.nan] * nrows, name="const_na")
data[3] = pd.Series(["s"] * nrows, name="const_str")
data[4] = pd.Series([0] * nrows, name="const_zero")
data[5] = pd.Series([-1] * nrows, name="const_neg")
data[6] = pd.Series([1] * nrows, name="const_pos")
data[7] = pd.Series([0, 1, np.nan] * (nrows // 3), name="small_distinct_miss")
data[8] = gen_random_series(size=nrows, dtype="string", random_state=rand).rename("str_no_miss")
data[9] = gen_random_series(size=nrows, dtype="string", na_ratio=0.1, random_state=rand).rename(
"str_miss"
)
data[10] = gen_random_series(size=nrows, dtype="float", random_state=rand).rename("num_no_miss")
data[11] = gen_random_series(size=nrows, dtype="float", na_ratio=0.1, random_state=rand).rename(
"num_miss"
)
data[12] = | pd.Series(["a", "b"] * (nrows // 2), name="category_no_miss", dtype="category") | pandas.Series |
# Preparation for Theme3 Cell of Origin using Panoptes
import pandas as pd
# train = pd.read_csv('../Theme3/train.csv', header=0)
# validation = pd.read_csv('../Theme3/val.csv', header=0)
# test = pd.read_csv('../Theme3/test.csv', header=0)
#
# cancer_dict = {'HNSCC': 0, 'CCRCC': 1, 'CO': 2, 'BRCA': 3, 'LUAD': 4, 'LSCC': 5, 'PDA': 6, 'UCEC': 7, 'GBM': 8, 'OV': 9}
#
# train['set'] = 'train'
# validation['set'] = 'validation'
# test['set'] = 'test'
#
# origin = pd.concat([train, validation, test])
# origin.columns = ['Tumor', 'Patient_ID', 'Slide_ID', 'set']
# origin['label'] = origin['Tumor'].replace(cancer_dict)
# pathls = []
# for idx, row in origin.iterrows():
# pathls.append("../tiles/{}/{}/{}/".format(str(row['Tumor']), str(row['Patient_ID']),
# row['Slide_ID'].split('-')[-1]))
# origin['path'] = pathls
#
# origin.to_csv('../Theme3_split.csv', index=False)
#
# DLCCA secondary prep
train = pd.read_csv('../DLCCA/train.csv', header=0)
validation = pd.read_csv('../DLCCA/val.csv', header=0)
test = | pd.read_csv('../DLCCA/test.csv', header=0) | pandas.read_csv |
from src.network import Network
import src.VisualizeNN as VisNN
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
def split_dataset(train_df: pd.DataFrame,
fraction: float = 0.2):
"""
Split train data into train and validation.
"""
#permute all samples
train_df = train_df.sample(frac=1.0)
#set validation fraction
fraction = 0.2
split_index = int(fraction*len(train_df))
valid_df = train_df.iloc[:split_index]
train_df = train_df.iloc[split_index:]
return train_df, valid_df
def get_normalisation_scale(df: pd.DataFrame,
regression: bool = False):
"""
Get normalization scales. Used later in normalize_dataset().
"""
if regression:
without_last_column = df
else:
# don't scale last column (class label)
without_last_column = df.iloc[:, :-1]
return (without_last_column.min(), without_last_column.max())
def normalized_dataset(df: pd.DataFrame,
df_min: pd.Series,
df_max: pd.Series,
regression: bool = False):
"""
Normalize dataset using calculated min and max values from get_normalisation_scale().
"""
# don't modify last column (class label)
if regression is False:
tmp_df = 2.0*((df.iloc[:,:-1]-df_min)/(df_max - df_min))-1.0
tmp_df['cls'] = df['cls']
else:
tmp_df = 2.0*((df-df_min)/(df_max - df_min))-1.0
return tmp_df
def train_classification( net: Network,
dataset: pd.DataFrame,
max_epochs: int,
learning_rate: float,
batch_size: int = 1,
multiclass: bool = False):
"""
Train net for a classification task.
The dataset consists of features and class label (in the last column).
If the multiclass is True, uses one-hot encoding.
"""
train_df, valid_df = split_dataset(dataset, 0.2)
train_y_df = pd.get_dummies(train_df['cls'], dtype=float) if multiclass else train_df['cls'] - 1.0
valid_y_df = | pd.get_dummies(valid_df['cls'], dtype=float) | pandas.get_dummies |
import os
import re
import pandas as pd
import metis_cut as metis
import kahip_cut as kahip
import inertialflow_cut as inertialflow
import flowcutter_cut as flowcutter
import inertialflowcutter_cut as ifc
experiments_folder = ""
graphs = ["col", "cal", "europe", "usa"]
partitioners = ["metis", "kahip_v2_11", "inertial_flow", "flowcutter3", "flowcutter20", "inertialflowcutter4", "inertialflowcutter8", "inertialflowcutter12", "inertialflowcutter16"]
imbalances = [0.0, 0.01, 0.03, 0.05, 0.1, 0.2, 0.3, 0.5, 0.7, 0.9]
binary_path = "./../build/"
console = binary_path + "console"
metis_path = "./" + experiments_folder + "gpmetis"
def graph_path(G):
return experiments_folder + G + "/"
def output_file(G, P):
return experiments_folder + G + "." + P + ".cut"
def compute_cuts(G, P):
if P == "metis":
return compute_metis_cuts(G)
elif P == "kahip_v2_11":
return compute_kahip_cuts(G, P, old=False)
elif P.startswith("flowcutter"):
cutters = int(re.match(r"flowcutter([0-9]+)", P).group(1))
return compute_flow_cutter_cuts(G, cutters)
elif P == 'inertial_flow':
return compute_inertial_flow_cuts(G)
elif P.startswith("inertialflowcutter"):
cutters = int(re.match(r"inertialflowcutter([0-9]+)", P).group(1))
return compute_inertial_flow_cutter_cuts(G, cutters)
else:
assert(false)
def compute_metis_cuts(G):
rows = []
for epsilon in imbalances:
row_dict = {}
metrics = metis.metis_cut(metis_path, console, graph_path(G), epsilon)
row_dict["epsilon"] = epsilon
row_dict["achieved_epsilon"] = metrics["epsilon"]
row_dict["cut_size"] = metrics["cut_size"]
row_dict["running_time"] = metrics["running_time"]
row_dict["connected"] = metrics["left_components"] == 1 and metrics["right_components"] == 1
rows.append(row_dict)
results = pd.DataFrame(rows)
return results.set_index("epsilon").sort_index()
def compute_kahip_cuts(G, P, old):
rows = []
for epsilon in imbalances:
row_dict = {}
metrics = kahip.kahip_cut(console, graph_path(G), epsilon)
row_dict["epsilon"] = epsilon
row_dict["achieved_epsilon"] = metrics["epsilon"]
row_dict["cut_size"] = metrics["cut_size"]
row_dict["running_time"] = metrics["running_time"]
row_dict["connected"] = metrics["left_components"] == 1 and metrics["right_components"] == 1
rows.append(row_dict)
results = | pd.DataFrame(rows) | pandas.DataFrame |
# -*- coding: utf-8 -*-
import pandas as pd
import numpy as np
import operator as op
import seaborn as sns
# http://data8.org/datascience/_modules/datascience/tables.html
#####################
# Frame Manipulation
def relabel(df, OriginalName, NewName):
return df.rename(index=str, columns={OriginalName: NewName})
# https://docs.python.org/3.4/library/operator.html
def where(df, column, value, operation=op.eq):
return pd.DataFrame( df.loc[operation(df.loc[:,column], value) ,:] )
def select(df, *column_or_columns):
table = pd.DataFrame()
for column in column_or_columns:
table[column] = df.loc[:, column].values
return table
def column(df, index_or_label):
"""Return the values of a column as an array.
Args:
label (int or str): The index or label of a column
Returns:
An instance of ``numpy.array``.
Raises:
``ValueError``: When the ``index_or_label`` is not in the table.
"""
if (isinstance(index_or_label, str)):
if (index_or_label not in df.columns):
raise ValueError(
'The column "{}" is not in the table. The table contains '
'these columns: {}'
.format(index_or_label, ', '.join(df.labels))
)
else:
return df.loc[:, index_or_label].values
if (isinstance(index_or_label, int)):
if (not 0 <= index_or_label < len(df.columns)):
raise ValueError(
'The index {} is not in the table. Only indices between '
'0 and {} are valid'
.format(index_or_label, len(df.labels) - 1)
)
else:
return df.iloc[:,index_or_label].values
def drop(df, index_or_label):
if (isinstance(index_or_label, str)):
if (index_or_label not in df.columns):
raise ValueError(
'The column "{}" is not in the table. The table contains '
'these columns: {}'
.format(index_or_label, ', '.join(df.labels))
)
else:
return df.drop(index_or_label, axis=1)
if (isinstance(index_or_label, int)):
if (not 0 <= index_or_label < len(df.columns)):
raise ValueError(
'The index {} is not in the table. Only indices between '
'0 and {} are valid'
.format(index_or_label, len(df.labels) - 1)
)
else:
return df.drop(index_or_label, axis=0)
return
def row(df, index):
"""Return the values of a row as an array.
Args:
label (int): The index or label of a column
Returns:
An instance of ``numpy.array``.
Raises:
``ValueError``: When the ``index_or_label`` is not in the table.
"""
return df.iloc[index,:].values
def cell(df, row, column):
return df.iloc[column, row]
def exclude(df, toexclude_df, column):
the_join = pd.merge(df, toexclude_df, on=[column], how="outer", indicator=True)
return ( pd.DataFrame(the_join).where('_merge', "left_only") )
def format(df, num_format=lambda x: '{:,.1f}'.format(x)):
"""Returns a better number formated table. Is Slow
Args:
label (int or str): The index or label of a column
Returns:
pandas dataframe
"""
#TODO: this looks inefficient
def build_formatters_ints(df):
return {
column:lambda x: '{:,}'.format(x)
for column, dtype in df.dtypes.items()
if dtype in [ np.dtype('int64') ]
}
def build_formatters_floats(df):
return {
column:lambda x: '{:.1f}'.format(x)
for column, dtype in df.dtypes.items()
if dtype in [ np.dtype('float64') ]
}
format_int = build_formatters_ints(df)
format_float = build_formatters_floats(df)
style = '<style>.dataframe td { text-align: right; }</style>'
return df.style.set_table_styles(style).format(format_int).format(format_float)
def group(df, column, rename=""):
df_gp = pd.DataFrame(df[column].value_counts())
if rename != "":
return relabel(df_gp,column,rename)
else:
return relabel(df_gp,column,column + "_count")
def count(df, column):
return len( np.unique( df[column] ))
def showna(df):
return sns.heatmap(df.isnull(),yticklabels=False,cbar=False,cmap='viridis')
def sort(df, col, ascending=True):
return pd.DataFrame(df.sort_values(col, ascending=ascending))
##
def variance(df, column1):
return np.var( pd.DataFrame(df)[column1] )
def median(df, column1):
return np.median( pd.DataFrame(df)[column1] )
def avg(df, column1):
return np.mean( | pd.DataFrame(df) | pandas.DataFrame |
# -*- coding: utf-8 -*-
"""
Created on Mon Jun 4 20:48:37 2018
@author: elcok
"""
import os
import sys
import numpy as np
import geopandas as gpd
import pandas as pd
sys.path.append(os.path.join( '..'))
from scripts.functions import region_exposure,region_losses,poly_files,load_sample
from scripts.utils import load_config,download_osm_file
import country_converter as coco
cc = coco.CountryConverter()
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
warnings.filterwarnings("ignore", category=DeprecationWarning)
pd.set_option('chained_assignment',None)
from multiprocessing import Pool,cpu_count
def all_countries_risk():
"""Function to estimate the risk for all countries consecutively.
"""
# specify country
countries = ['LU','CZ','CH','EE','LV','LT','PT','ES','AT','BE','DK','IE','NL','NO','SE','UK','PL','IT','FI','FR','DE']
for country in countries:
losses(country, parallel = False, event_set = True)
def all_countries_losses():
"""Function to estimate the losses for all countries consecutively.
"""
# specify country
countries = ['LU','CZ','CH','EE','LV','LT','PT','ES','AT','BE','DK','IE','NL','NO','SE','UK','PL','IT','FI','FR','DE']
for country in countries:
losses(country, parallel = True)
def all_countries_exposure():
"""Function to estimate the exposure for all countries consecutively.
"""
# specify country
countries = ['LU','CZ','CH','EE','LV','LT','PT','ES','AT','BE','DK','IE','NL','NO','SE','UK','PL','IT','FI','FR','DE']
for country in countries:
exposure(country, include_storms = True, parallel = False)
def exposure(country, include_storms = True, parallel = True,save=True):
"""
Creation of exposure table of the specified country.
Arguments:
*country* (string) -- ISO2 code of country to consider.
*include_storms* (bool) -- if set to False, it will only return a list of buildings and their characteristics (default: **True**).
*parallel* (bool) -- calculates all regions within a country parallel. Set to False if you have little capacity on the machine (default: **True**).
*save* (bool) -- boolean to decide whether you want to save the output to a csv file (default: **True**).
Returns:
*GeoDataframe* -- Geopandas dataframe with all buildings of the country and potential exposure to wind
"""
#make sure the country inserted is an ISO2 country name for he remainder of the analysis
#country = coco.convert(names=country, to='ISO2')
# get data path
data_path = load_config()['paths']['data']
# create country poly files
poly_files(data_path,country)
#download OSM file if it is not there yet:
download_osm_file(country)
#get list of regions for which we have poly files (should be all)
regions = os.listdir(os.path.join(data_path,country,'NUTS3_POLY'))
regions = [x.split('.')[0] for x in regions]
if include_storms == True:
storms = len(regions)*[True]
country_list = len(regions)*[country]
if parallel == True:
with Pool(cpu_count()-2) as pool:
country_table = pool.starmap(region_exposure,zip(regions,country_list,storms),chunksize=1)
else:
country_table = []
for region in regions:
country_table.append(region_exposure(region,country,True))
else:
storms = len(regions)*[False]
country_list = len(regions)*[country]
if parallel == True:
with Pool(cpu_count()-2) as pool:
country_table = pool.starmap(region_exposure,zip(regions,country_list,storms),chunksize=1)
else:
country_table = []
for region in regions:
country_table.append(region_exposure(region,country,True))
if save == True:
gdf_table = gpd.GeoDataFrame(pd.concat(country_table),crs='epsg:4326')
gdf_table.drop(['centroid'],axis='columns',inplace=True)
gdf_table.to_file(os.path.join(data_path,'exposure_country',country,'{}_exposure.shp'.format(country)))
return gpd.GeoDataFrame(pd.concat(country_table),crs='epsg:4326')
def losses(country, parallel = True, event_set = False,save=True):
"""
Creation of exposure table of the specified country
Arguments:
*country* (string) -- ISO2 code of country to consider.
*parallel* (bool) -- calculates all regions within a country parallel. Set to False if you have little capacity on the machine (default: **True**).
*event_set* (bool) -- if set to True, we will calculate the losses for the event set instead of the historical storms (default: **True**).
*save* (bool) -- boolean to decide whether you want to save the output to a csv file (default: **True**).
Returns:
*GeoDataframe* -- Geopandas dataframe with all buildings of the country and their **losses** for each wind storm.
"""
#make sure the country inserted is an ISO2 country name for he remainder of the analysis
#country = coco.convert(names=country, to='ISO2')
# get data path
data_path = load_config()['paths']['data']
# create country poly files
poly_files(data_path,country)
#download OSM file if it is not there yet:
download_osm_file(country)
#load sample
sample = load_sample(country)
#get list of regions for which we have poly files (should be all)
regions = os.listdir(os.path.join(data_path,country,'NUTS3_POLY'))
regions = [x.split('.')[0] for x in regions]
if event_set == False:
event_set = len(regions)*[False]
samples = len(regions)*[sample]
if parallel == True:
with Pool(cpu_count()-2) as pool:
country_table = pool.starmap(region_losses,zip(regions,event_set,samples),chunksize=1)
else:
country_table = []
for region in regions:
country_table.append(region_losses(region,False,sample))
elif event_set == True:
event_set = len(regions)*[True]
samples = len(regions)*[sample]
if parallel == True:
with Pool(cpu_count()-2) as pool:
country_table = pool.starmap(region_losses,zip(regions,event_set,samples),chunksize=1)
else:
country_table = []
for region in regions:
country_table.append(region_losses(region,True))
if (save == True) & (event_set == False):
gdf_table = gpd.GeoDataFrame( | pd.concat(country_table) | pandas.concat |
import numpy as np
import random
from flask import Flask, request, render_template
from model.simple_recommender_model import simple_recommend
import pandas as pd
from tensorflow import keras
from rake_nltk import Rake
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.feature_extraction.text import CountVectorizer
app = Flask(__name__)
#<EMAIL>('/')
#def homepage():
# return render_template('homepage.html')
#<EMAIL>('/watchtrailer')
#def watch():
# return render_template('fresh_tomatoes.html')
@app.route('/', methods=['GET', 'POST'])
def form_example():
if request.method == 'POST':
moviesall=[]
movies=[]
def xstr(s):
if s is None:
return ""
else:
return str(s)
Movie1 = request.form.get('chk1')
Movie1 = xstr(Movie1)
moviesall.append(Movie1)
Movie2 = request.form.get('chk2')
Movie2 = xstr(Movie2)
moviesall.append(Movie2)
Movie3 = request.form.get('chk3')
Movie3 = xstr(Movie3)
moviesall.append(Movie3)
Movie4 = request.form.get('chk4')
Movie4 = xstr(Movie4)
moviesall.append(Movie4)
Movie5 = request.form.get('chk5')
Movie5 = xstr(Movie5)
moviesall.append(Movie5)
Movie6 = request.form.get('chk6')
Movie6 = xstr(Movie6)
moviesall.append(Movie6)
Movie7 = request.form.get('chk7')
Movie7 = xstr(Movie7)
moviesall.append(Movie7)
Movie8 = request.form.get('chk8')
Movie8 = xstr(Movie8)
moviesall.append(Movie8)
Movie9 = request.form.get('chk9')
Movie9 = xstr(Movie9)
moviesall.append(Movie9)
Movie10 = request.form.get('chk10')
Movie10 = xstr(Movie10)
moviesall.append(Movie10)
Movie11 = request.form.get('chk11')
Movie11 = xstr(Movie11)
moviesall.append(Movie11)
Movie12 = request.form.get('chk12')
Movie12 = xstr(Movie12)
moviesall.append(Movie12)
Movie13 = request.form.get('chk13')
Movie13 = xstr(Movie13)
moviesall.append(Movie13)
Movie14 = request.form.get('chk14')
Movie14 = xstr(Movie14)
moviesall.append(Movie14)
Movie15 = request.form.get('chk15')
Movie15 = xstr(Movie15)
moviesall.append(Movie15)
Movie16 = request.form.get('chk16')
Movie16 = xstr(Movie16)
moviesall.append(Movie16)
Movie17 = request.form.get('chk17')
Movie17 = xstr(Movie17)
moviesall.append(Movie17)
Movie18 = request.form.get('chk18')
Movie18 = xstr(Movie18)
moviesall.append(Movie18)
Movie19 = request.form.get('chk19')
Movie19 = xstr(Movie19)
moviesall.append(Movie19)
Movie20 = request.form.get('chk20')
Movie20 = xstr(Movie20)
moviesall.append(Movie20)
Movie21 = request.form.get('chk21')
Movie21 = xstr(Movie21)
moviesall.append(Movie21)
Movie22 = request.form.get('chk22')
Movie22 = xstr(Movie22)
moviesall.append(Movie22)
Movie23 = request.form.get('chk23')
Movie23 = xstr(Movie23)
moviesall.append(Movie23)
Movie24 = request.form.get('chk24')
Movie24 = xstr(Movie24)
moviesall.append(Movie24)
Movie25 = request.form.get('chk25')
Movie25 = xstr(Movie25)
moviesall.append(Movie25)
Movie26 = request.form.get('chk26')
Movie26 = xstr(Movie26)
moviesall.append(Movie26)
Movie27 = request.form.get('chk27')
Movie27 = xstr(Movie27)
moviesall.append(Movie27)
Movie28 = request.form.get('chk28')
Movie28 = xstr(Movie28)
moviesall.append(Movie28)
Movie29 = request.form.get('chk29')
Movie29 = xstr(Movie29)
moviesall.append(Movie29)
Movie30 = request.form.get('chk30')
Movie30 = xstr(Movie30)
moviesall.append(Movie30)
Movie31 = request.form.get('chk31')
Movie31 = xstr(Movie31)
moviesall.append(Movie31)
Movie32 = request.form.get('chk32')
Movie32 = xstr(Movie32)
moviesall.append(Movie32)
Movie33 = request.form.get('chk33')
Movie33 = xstr(Movie33)
moviesall.append(Movie33)
Movie34 = request.form.get('chk34')
Movie34 = xstr(Movie34)
moviesall.append(Movie34)
Movie35 = request.form.get('chk35')
Movie35 = xstr(Movie35)
moviesall.append(Movie35)
Movie36 = request.form.get('chk36')
Movie36 = xstr(Movie36)
moviesall.append(Movie36)
for item in moviesall:
if item:
movies.append(item)
else:
continue
dataset = pd.read_csv(r'C:\Users\Lenovo PC\Downloads\ml-latest-small\ratings.csv')
M = | pd.read_csv(r'C:\Users\Lenovo PC\Desktop\Summary Work - Sheet1.csv', encoding='latin-1') | pandas.read_csv |
import pandas as pd
import matplotlib.pyplot as plt
import scipy.stats
from sklearn import linear_model
import statsmodels.api as sm
from scipy import stats
###################
yaara="723"
daniel = "957"
hilla="355"
generic_path = "/tmp/pycharm_project_"+hilla+"/"
#Full data
dfOp = pd.read_csv("/mnt/nadavrap-students/STS/data/Shapira_1st-Op_6_9_20_.csv")
groupOp = dfOp.groupby("SiteID")["SiteID"].count().reset_index(name='countFirst')
#draw a plot
x = groupOp["countFirst"]
plt.hist(x, bins=40)
plt.title("Histogram of count Operation")
plt.xlabel('number of Operations')
plt.ylabel('count of SiteId')
plt.show()
plt.savefig('Histogram of count Operation.png')
#ReOp data
dfReOp = pd.read_csv("/mnt/nadavrap-students/STS/data/Shapira_reOp_6_9_20_.csv")
groupReOp = dfReOp.groupby("SiteID")["SiteID"].count().reset_index(name='countReOp')
#draw a plot
y = groupReOp['countReOp']
plt.hist(y, bins=20)
plt.title("Histogram of count ReOperation")
plt.xlabel('number of ReOperations')
plt.ylabel('count of SiteId')
plt.show()
plt.savefig('Histogram of count ReOperation.png')
##merge two dataframes into one and gets the ratio between them
result = pd.merge(groupOp, groupReOp, on='SiteID', how='left')
result['countReOp'].fillna(0, inplace=True)
result["countReOp/countFirst+countReOp"] = (result["countReOp"] /(result["countReOp"]+ result["countFirst"])) *100
result['countReOp/countFirst+countReOp'].fillna(0, inplace=True)
result.to_csv(generic_path+"result.csv")
#draw a plot
z = result['countReOp/countFirst+countReOp']
plt.hist(z, bins=40)
plt.title("Histogram of ReOperation vs Operation")
plt.xlabel('% of ReOperation of Operation')
plt.ylabel('count of SiteId')
plt.show()
plt.savefig('Histogram of ReOperation vs Operation.png')
########### nadav recomend ###############
# import feather
# feather.write_dataframe(dfOp, "/tmp/pycharm_project_723/dfOp.feather")
# feather.write_dataframe(dfReOp, "/tmp/pycharm_project_723/dfReOp.feather")
# dfop1 = feather.read_dataframe("/tmp/pycharm_project_723/dfOp.feather")
# dfReOp1 = feather.read_dataframe("/tmp/pycharm_project_723/dfReOp.feather")
######mortality
MortaltyOp = dfOp.groupby('SiteID')['Mortalty'].apply(lambda x: (x== 1 ).sum()).reset_index(name='Mortalty_SiteID_op')
MortaltyReOp = dfReOp.groupby('SiteID')['Mortalty'].apply(lambda x: (x== 1 ).sum()).reset_index(name='Mortalty_SiteID_reOp')
result2 = pd.merge(MortaltyOp, MortaltyReOp, on='SiteID', how='left')
# result.merge(result2, on='SiteID')
df=pd.merge(result, result2, on='SiteID')
df["countOpr"] = result["countReOp"]+ result["countFirst"]
countOpr=df['countOpr']
df.to_csv(generic_path+"mortalty.csv")
####AGE
ageOp = dfOp.groupby("SiteID")["Age"].mean().reset_index(name='Mean_Age_op')
ageReOp = dfReOp.groupby("SiteID")["Age"].mean().reset_index(name='Mean_Age_reOp')
resultAge = pd.merge(ageOp, ageReOp, on='SiteID', how='left')
dfAge=pd.merge(result, resultAge, on='SiteID')
genderOp = | pd.get_dummies(dfOp["Gender"]) | pandas.get_dummies |
import urllib
import pytest
import pandas as pd
from pandas import testing as pdt
from anonympy import __version__
from anonympy.pandas import dfAnonymizer
from anonympy.pandas.utils_pandas import load_dataset
@pytest.fixture(scope="module")
def anonym_small():
df = load_dataset('small')
anonym = dfAnonymizer(df)
return anonym
@pytest.fixture(scope="module")
def anonym_big():
try:
df = load_dataset('big')
anonym = dfAnonymizer(df)
except urllib.error.HTTPError:
anonym = None
return anonym
def test_anonym_obj(anonym_small, anonym_big):
assert isinstance(anonym_small, dfAnonymizer), "should have\
returned `dfAnonymizer` object"
if anonym_big is None:
assert False, "Failed to fetch the DataFrame"
assert isinstance(anonym_big, dfAnonymizer), "should have returned\
`dfAnonymizer` object"
def test_numeric_noise(anonym_small):
output = anonym_small.numeric_noise('age', seed=42, inplace=False)
expected = pd.Series([38, 47], dtype='int64')
pdt.assert_series_equal(expected, output, check_names=False)
output = anonym_small.numeric_noise(['age', 'salary'],
seed=42,
inplace=False)
expected = pd.DataFrame({'age': [38, 47],
'salary': [59239.79912097112, 49323.30756879504]})
pdt.assert_frame_equal(expected, output)
def test_numeric_binning(anonym_small):
output = anonym_small.numeric_binning('salary', bins=2, inplace=False)
dtype = pd.CategoricalDtype([
pd.Interval(49315.0, 54279.0, closed='right'),
pd.Interval(54279.0, 59234.0, closed='right')],
ordered=True)
expected = pd.Series([
pd.Interval(54279.0, 59234.0, closed='right'),
pd.Interval(49315.0, 54279.0, closed='right')],
dtype=dtype)
pdt.assert_series_equal(expected, output, check_names=False)
output = anonym_small.numeric_binning(['age', 'salary'],
bins=2,
inplace=False)
dtype2 = pd.CategoricalDtype([
pd.Interval(33.0, 40.0, closed='right'),
pd.Interval(40.0, 48.0, closed='right')],
ordered=True)
ser2 = pd.Series([
pd.Interval(33.0, 40.0, closed='right'),
pd.Interval(40.0, 48.0, closed='right')],
dtype=dtype2)
expected = pd.DataFrame({'age': ser2, 'salary': expected})
pdt.assert_frame_equal(expected, output)
def test_numeric_masking(anonym_small):
output = anonym_small.numeric_masking('age', inplace=False)
expected = pd.Series([7.5, -7.5], dtype='float64')
pdt.assert_series_equal(expected, output, check_names=False)
output = anonym_small.numeric_masking(['age', 'salary'], inplace=False)
expected = pd.DataFrame({'age': [-4954.900676201789, 4954.900676201798],
'salary': [5.840670901327418e-15,
5.840670901327409e-15]})
pdt.assert_frame_equal(expected, output)
def test_numeric_rounding(anonym_small):
output = anonym_small.numeric_rounding('salary', inplace=False)
expected = pd.Series([60000.0, 50000.0], dtype='float64')
pdt.assert_series_equal(expected, output, check_names=False)
output = anonym_small.numeric_rounding(['age', 'salary'], inplace=False)
expected = pd.DataFrame({'age': {0: 30, 1: 50}, 'salary': {0: 60000.0,
1: 50000.0}})
| pdt.assert_frame_equal(expected, output) | pandas.testing.assert_frame_equal |
# %%%%
import pandas as pd
import numpy as np
import re
# %%%% functions
## Fill missing values
def fillmissing(x,col,index,benchmark):
for i in range(index,len(x)):
# find missing value
if x.loc[i,col] == benchmark:
# if first is missing, fill using the value next to it
if i == index:
x.loc[i,col] = x.loc[i+1,col]
# if the last one is missing, fill using the value preceeds it
elif i == len(x)-1:
x.loc[i,col] = x.loc[i-1,col]
# otherwise, fill using the average of the two not null values above and after
else:
j = i-1
k = i+1
while x.loc[j,col] == benchmark:
j -= 1
while x.loc[k,col] == benchmark:
k += 1
x.loc[i,col] = np.mean([x.loc[j,col],x.loc[k,col]])
return x
## Data Preprocess
def preprocess(x,name,Date,column,index,benchmark,q):
# select the valid starting day
x = x[x['Date'] > Date].copy()
x = x.reset_index().copy()
x = x.drop('index',axis = 1).copy()
# fill na with benchmark we chose
x[column] = x[column].fillna(benchmark).copy()
# fill missing values
x = fillmissing(x,column,index,benchmark).copy()
# calculate daily return
x['lag_'+column] = x[column].shift(1)
x = x.iloc[1:,:].copy().reset_index()
x = x.drop('index',axis = 1).copy()
x['log_ret'] = np.log(x[column])-np.log(x['lag_'+column])
retm = np.mean(x['log_ret'])
x['retv'] = np.square(x['log_ret']-retm)*100
# estimate volatility
x[name+'_20day_vol'] = np.sqrt(x['retv'].rolling(window=20,win_type="boxcar").mean())/10
# estimate quantiles of the distribution of log-returns
x[name+'_quant_ret'] = np.nan
for r in range(len(x)-20):
R_quant = np.quantile(x['log_ret'][r:r+20],q)
x.loc[r+19,name+'_quant_ret'] = R_quant
return x
# %%%% Main Dataset: csi300
csi = pd.read_csv('/Users/msstark/Desktop/project/Shanghai Shenzhen CSI 300 Historical Data.csv')
# setting date format
csi['Date'] = csi['Date'].apply(lambda x: re.sub(r',',r'',x))
csi['Day'] = csi['Date'].apply(lambda x: x.split(' ')[1]).astype(int)
csi['Month'] = csi['Date'].apply(lambda x: x.split(' ')[0])
csi['Month'].unique()
csi['Month'] = csi['Month'].map({'Jan':1,'Feb':2,'Mar':3,'Apr':4,'May':5,'Jun':6,
'Jul':7,'Aug':8,'Sep':9,'Oct':10,'Nov':11,'Dec':12})
csi['Year'] = csi['Date'].apply(lambda x: x.split(' ')[2]).astype(int)
csi['Date'] = csi['Year'].astype(str) +'-'+csi['Month'].astype(str)+'-'+csi['Day'].astype(str)
csi['Date'] = pd.to_datetime(csi['Date'], format='%Y-%m-%d')
csi = csi.rename(columns = {'Price':'Close'}).copy()
# convert object type to float
col = ['Close','Open','High','Low']
for c in col:
csi[c] = csi[c].apply(lambda x: re.sub(r',',r'',x)).astype('float')
csi['log_dsprd'] = np.log(csi['High'] - csi['Low'])
csi.columns
# apply preprocess function
csi = preprocess(csi,'csi','2005-01-03','Close',0,0,0.025).copy()
# %%%% spot exchange rate
xr = pd.read_csv('/Users/msstark/Desktop/project/DEXCHUS.csv')
# setting date format
xr['DATE'] = | pd.to_datetime(xr['DATE'], format='%Y-%m-%d') | pandas.to_datetime |
import pymortar
import pandas as pd
import pendulum
import toml
from flask import Flask
from flask import jsonify, send_from_directory
from flask import request
from flask import current_app
from flask import make_response
from flask import render_template
from collections import defaultdict
from functools import update_wrapper
import pytz
import json
import glob
import os
from datetime import datetime, timedelta
from dashutil import get_start, generate_months, prevmonday, get_today
from datetime import timezone
import xsg
config = toml.load('config.toml')
TZ = pytz.timezone('US/Pacific')
app = Flask(__name__, static_url_path='/static')
client = pymortar.Client({
'mortar_address':config['Mortar']['url'],
'username': config['Mortar']['username'],
'password': config['<PASSWORD>']['password'],
})
sites = [config['Dashboard']['sitename']]
def crossdomain(origin=None, methods=None, headers=None,
max_age=21600, attach_to_all=True,
automatic_options=True):
if methods is not None:
methods = ', '.join(sorted(x.upper() for x in methods))
if headers is not None and not isinstance(headers, str):
headers = ', '.join(x.upper() for x in headers)
if not isinstance(origin, str):
origin = ', '.join(origin)
if isinstance(max_age, timedelta):
max_age = max_age.total_seconds()
def get_methods():
if methods is not None:
return methods
options_resp = current_app.make_default_options_response()
return options_resp.headers['allow']
def decorator(f):
def wrapped_function(*args, **kwargs):
if automatic_options and request.method == 'OPTIONS':
resp = current_app.make_default_options_response()
else:
resp = make_response(f(*args, **kwargs))
if not attach_to_all and request.method != 'OPTIONS':
return resp
h = resp.headers
h['Access-Control-Allow-Origin'] = origin
h['Access-Control-Allow-Methods'] = get_methods()
h['Access-Control-Max-Age'] = str(max_age)
if headers is not None:
h['Access-Control-Allow-Headers'] = headers
return resp
f.provide_automatic_options = False
return update_wrapper(wrapped_function, f)
return decorator
def state_to_string(state):
if state == 0:
return 'off'
elif state == 1:
return 'heat stage 1'
elif state == 2:
return 'cool stage 1'
elif state == 4:
return 'heat stage 2'
elif state == 5:
return 'cool stage 2'
else:
return 'unknown'
def dofetch(views, dataframes, start=None, end=None):
timeparams = None
if start is not None and end is not None:
timeparams=pymortar.TimeParams(
start=start.isoformat(),
end=end.isoformat(),
)
req = pymortar.FetchRequest(
sites=sites,
views=views,
dataFrames=dataframes,
time=timeparams
)
return client.fetch(req)
meter_view = pymortar.View(
name="meters",
definition="""SELECT ?meter WHERE {
?meter rdf:type brick:Building_Electric_Meter
};""",
)
meter_df = pymortar.DataFrame(
name="meters",
aggregation=pymortar.MEAN,
timeseries=[
pymortar.Timeseries(
view="meters",
dataVars=['?meter'],
)
]
)
tstats_view = pymortar.View(
name="tstats",
definition="""SELECT ?rtu ?zone ?tstat ?csp ?hsp ?temp ?state WHERE {
?rtu rdf:type brick:RTU .
?tstat bf:controls ?rtu .
?rtu bf:feeds ?zone .
?tstat bf:hasPoint ?temp .
?temp rdf:type/rdfs:subClassOf* brick:Temperature_Sensor .
?tstat bf:hasPoint ?csp .
?csp rdf:type/rdfs:subClassOf* brick:Supply_Air_Temperature_Heating_Setpoint .
?tstat bf:hasPoint ?hsp .
?hsp rdf:type/rdfs:subClassOf* brick:Supply_Air_Temperature_Cooling_Setpoint .
?tstat bf:hasPoint ?state .
?state rdf:type brick:Thermostat_Status .
};""",
)
tstats_df = pymortar.DataFrame(
name="tstats",
aggregation=pymortar.MAX,
timeseries=[
pymortar.Timeseries(
view="tstats",
dataVars=['?csp','?hsp','?temp','?state'],
),
]
)
room_temp_view = pymortar.View(
name="room_temp",
definition="""SELECT ?zone ?room ?sensor WHERE {
?zone rdf:type brick:HVAC_Zone .
?zone bf:hasPart ?room .
?sensor rdf:type/rdfs:subClassOf* brick:Temperature_Sensor .
?room bf:hasPoint ?sensor .
};""",
)
weather_view = pymortar.View(
name="weather_temp",
definition="""SELECT ?sensor WHERE {
?sensor rdf:type/rdfs:subClassOf* brick:Weather_Temperature_Sensor .
};""",
)
weather_df = pymortar.DataFrame(
name="weather_temp",
aggregation=pymortar.MEAN,
window='15m',
timeseries=[
pymortar.Timeseries(
view="weather_temp",
dataVars=['?sensor'],
)
],
)
@app.route('/api/power/<last>/in/<bucketsize>')
@crossdomain(origin='*')
def power_summary(last, bucketsize):
# first, determine the start date from the 'last' argument
start_date = get_start(last)
if last == 'year' and bucketsize == 'month':
ranges = generate_months(get_today().month - 1)
readings = []
times = []
for t0,t1 in ranges:
meter_df.window = '{0}d'.format((t0-t1).days)
res=dofetch([meter_view], [meter_df], t1, t0)
times.append(t1.tz_convert(TZ).timestamp()*1000)
readings.append(res['meters'].fillna('myNullVal').values[0][0])
return jsonify({'readings': dict(zip(times,readings))})
# otherwise,
meter_df.window=bucketsize
print('start_date',start_date)
res=dofetch([meter_view], [meter_df], start_date, datetime.now(TZ))
res['meters'].columns=['readings']
return res['meters'].tz_convert(TZ).fillna('myNullVal').to_json()
@app.route('/api/energy/<last>/in/<bucketsize>')
@crossdomain(origin='*')
def energy_summary(last, bucketsize):
start_date = get_start(last)
if last == 'year' and bucketsize == 'month':
ranges = generate_months(get_today().month - 1)
readings = []
times = []
for t0,t1 in ranges:
meter_df.window = '15m'
res=dofetch([meter_view], [meter_df], t1, t0)
df = res['meters'].copy()
df.columns = ['readings']
df /= 4. # divide by 4 to get 15min (kW) -> kWh
times.append(pd.to_datetime(t1.isoformat()))
readings.append(df['readings'].sum())
df = | pd.DataFrame(readings,index=times,columns=['readings']) | pandas.DataFrame |
""" test get/set & misc """
from datetime import timedelta
import re
import numpy as np
import pytest
from pandas import (
DataFrame,
IndexSlice,
MultiIndex,
Series,
Timedelta,
Timestamp,
date_range,
period_range,
timedelta_range,
)
import pandas._testing as tm
def test_basic_indexing():
s = Series(np.random.randn(5), index=["a", "b", "a", "a", "b"])
msg = "index 5 is out of bounds for axis 0 with size 5"
with pytest.raises(IndexError, match=msg):
s[5]
with pytest.raises(IndexError, match=msg):
s[5] = 0
with pytest.raises(KeyError, match=r"^'c'$"):
s["c"]
s = s.sort_index()
with pytest.raises(IndexError, match=msg):
s[5]
msg = r"index 5 is out of bounds for axis (0|1) with size 5|^5$"
with pytest.raises(IndexError, match=msg):
s[5] = 0
def test_basic_getitem_with_labels(datetime_series):
indices = datetime_series.index[[5, 10, 15]]
result = datetime_series[indices]
expected = datetime_series.reindex(indices)
tm.assert_series_equal(result, expected)
result = datetime_series[indices[0] : indices[2]]
expected = datetime_series.loc[indices[0] : indices[2]]
tm.assert_series_equal(result, expected)
def test_basic_getitem_dt64tz_values():
# GH12089
# with tz for values
ser = Series(
date_range("2011-01-01", periods=3, tz="US/Eastern"), index=["a", "b", "c"]
)
expected = Timestamp("2011-01-01", tz="US/Eastern")
result = ser.loc["a"]
assert result == expected
result = ser.iloc[0]
assert result == expected
result = ser["a"]
assert result == expected
def test_getitem_setitem_ellipsis():
s = Series(np.random.randn(10))
np.fix(s)
result = s[...]
tm.assert_series_equal(result, s)
s[...] = 5
assert (result == 5).all()
@pytest.mark.parametrize(
"result_1, duplicate_item, expected_1",
[
[
Series({1: 12, 2: [1, 2, 2, 3]}),
Series({1: 313}),
Series({1: 12}, dtype=object),
],
[
| Series({1: [1, 2, 3], 2: [1, 2, 2, 3]}) | pandas.Series |
from time import sleep
from os import getcwd, makedirs
from datetime import datetime
from pandas import DataFrame, concat
from config.config import CONFIG
from thread_runner.runner import ThreadRunner
from models.house import House
from models.room import Room
from models.datetime import Datetime
COLUMNS = [
'timestamp',
'currentTemperature',
'targetTemperature',
'backyardTemperature',
'heaterPower',
'maxHeaterPower',
'numberOfPeople'
]
class CSVLogger(ThreadRunner):
def __init__(self, logging_interval: float, house: House, datetime_: Datetime):
super().__init__()
self.enabled = CONFIG.get('csvLoggerEnabled', False)
root_path = f'{getcwd()}'
self.logs_path = f'{root_path}/logs'
self.logging_interval = logging_interval
self.house = house
self.datetime = datetime_
self.room_dataframes = self.__get_prepared_room_dataframes()
self.__create_directory(self.logs_path)
def _run(self):
log_directory = self.__init_logging_process()
while self.is_running:
for room_id, dataframe in self.room_dataframes.items():
room = self.house.get_room_by_id(room_id)
latest_data = self.__get_latest_data(room)
self.room_dataframes[room_id] = | concat((self.room_dataframes[room_id], latest_data), ignore_index=True) | pandas.concat |
# import pandas and numpy, and load the nls data
import pandas as pd
pd.set_option('display.width', 80)
| pd.set_option('display.max_columns', 7) | pandas.set_option |
import ast
import os
import pandas as pd
data_folder = "../../data"
num_of_workpiece = 20
workpiece_list = [f"wp_{idx + 1}" for idx in range(num_of_workpiece)]
workcell_list = [f"wc_{idx + 1}" for idx in range(17)]
input_filename = "input_3"
score_type = "independent_qc"
threshold = 0.9
input_folder = f"{data_folder}/raw/{input_filename}/independent_qc/{score_type}/{num_of_workpiece}_workpiece"
output_folder = f"{data_folder}/postprocessed/independent_qc/{score_type}/{num_of_workpiece}_workpiece/per_input"
if not os.path.exists(output_folder):
# Create target Directory
os.makedirs(output_folder)
print("Directory ", output_folder, " Created ")
else:
print("Directory ", output_folder, " already exists")
writer = pd.ExcelWriter(f"{output_folder}/independent_qc_{score_type}_{num_of_workpiece}_workpiece_{input_filename}.xlsx", mode="w")
for idx, folder in enumerate(os.listdir(f"{input_folder}")): # Each datetime folder
print(folder)
data = {
"threshold": [],
"qc_iteration": [], # 1 iteration = start of inspection until reach consent
"workpiece": [],
"num_of_winner_qc": [],
"MSE": [],
}
for i in range(17):
data[f"wc_{i + 1}_score"] = []
data[f"wc_{i + 1}_capability"] = []
compiled_df = pd.DataFrame(data)
# Fill in score and capability of first row
filename = os.listdir(f"{input_folder}/{folder}")[0] # Using first filename
df = pd.read_csv(f"{input_folder}/{folder}/{filename}")
capability_dict = dict()
for workcell in workcell_list:
data[f"{workcell}_score"] = 0.1
data[f"{workcell}_capability"] = df[df["qc"] == workcell].qc_capability.iloc[0]
# Concatenate consent_df and reset index to get iteration number
df_list = list()
for workpiece in workpiece_list:
for filename in os.listdir(f"{input_folder}/{folder}"): # Each results
if f"{workpiece}_" in filename:
# print(filename)
suffix = f"_{threshold}.csv"
if filename.endswith(suffix):
df = | pd.read_csv(f"{input_folder}/{folder}/{filename}") | pandas.read_csv |
#!/usr/bin/env python
import os
import logging
import argparse
import numpy as np
import pandas as pd
import logging.handlers
from .__init__ import __version__
from .plot import make_color_dict, plot_legend, plot_passages, plot_appearance
from .colorlog import ColorFormatter
logger = logging.getLogger('evol')
def set_logging(v):
logger.propagate = True
logger.setLevel(logging.DEBUG)
ch = logging.StreamHandler()
if v == 0:
ch.setLevel(logging.INFO)
elif v >= 1:
ch.setLevel(logging.DEBUG)
formatter = ColorFormatter('%(asctime)s - %(name)s - $COLOR%(message)s$RESET','%H:%M:%S')
ch.setFormatter(formatter)
logger.addHandler(ch)
def get_options():
description = 'Plot OD600 values for an evolution experiment'
parser = argparse.ArgumentParser(description=description)
parser.add_argument('data',
nargs='+',
help='Input reading from plate reader; '
'should contain the following columns: '
'"row", "column", '
'"plate", "passage", "strain", "treatment", '
'"concentration", '
'"experiment", "od600"')
parser.add_argument('output',
help='Output directory')
parser.add_argument('--threshold',
type=float,
default=0.5,
help='OD600 threshold to call growth '
'(default: %(default).2f)')
parser.add_argument('--format',
choices=('png',
'tiff',
'pdf',
'svg'),
default='png',
help='Output format (default: %(default)s)')
parser.add_argument('-v', action='count',
default=0,
help='Increase verbosity level')
parser.add_argument('--version', action='version',
version='%(prog)s '+__version__)
return parser.parse_args()
def main():
options = get_options()
set_logging(options.v)
df = []
for filename in options.data:
logger.info(f'reading data from {filename}')
df.append(pd.read_csv(filename, sep='\t'))
df = pd.concat(df)
# make sure we don't group replicates together
df['id'] = [f'{e}{p}{x}{y}'
for e, p, x, y in
df[['experiment', 'plate', 'row', 'column']].values]
# drop data with no treatment
df = df.loc[df['treatment'].dropna().index].copy()
logger.info(f'plotting strains legend')
strains = sorted(set(df['strain'].dropna().unique()
).difference(['Media Control']))
strains_colors = make_color_dict(strains, cmap='hsv')
fname = os.path.join(options.output, f'strain_legend.{options.format}')
plot_legend(strains_colors, fname)
df['treatment-id'] = [f'{treatment}-{conc}'
if treatment != 'GC'
else 'GC'
for treatment, conc in df[['treatment',
'concentration']].values]
logger.info(f'plotting treatments legend')
treatments = sorted(set(df['treatment-id'].dropna().unique()))
treatments_colors = make_color_dict(treatments, cmap='tab10')
fname = os.path.join(options.output, f'treatment_legend.{options.format}')
plot_legend(treatments_colors, fname)
# pivot the tables
op = df.pivot_table(index=['treatment-id', 'strain', 'id'],
columns=['passage'], values='od600')
logger.info(f'plotting all passages')
fname = os.path.join(options.output, f'passages.{options.format}')
plot_passages(op, treatments_colors, strains_colors, fname, 'OD600')
# pivot the tables (average)
op = df.pivot_table(index=['treatment-id', 'strain'],
columns=['passage'], values='od600')
logger.info(f'plotting all passages (average)')
fname = os.path.join(options.output, f'passages_average.{options.format}')
plot_passages(op, treatments_colors, strains_colors, fname, 'OD600 (average)')
# first appearance
logger.info(f'computing the first appearance of resistance')
# might fail, deal with errors gracefully
try:
df = df[df['passage'] > 0].copy()
appearance = []
for _, x in df.iterrows():
if x['od600'] < options.threshold:
v = 0
else:
if x['passage'] == df[df['treatment-id'] == df['treatment-id']]['passage'].max():
v = 1
elif df[(df['id'] == x['id']) &
(df['treatment'] == x['treatment']) &
(df['strain'] == x['strain']) &
(df['passage'] == x['passage'] + 1)]['od600'].values[0] >= options.threshold:
v = 1
else:
v = 0
appearance.append(v)
df['appearance'] = appearance
app = df[df['appearance'] > 0].groupby([
'treatment-id', 'strain', 'id'])['passage'].min().reset_index()
no_app = df[df['appearance'] == 0].groupby([
'treatment-id', 'strain', 'id'])['passage'].max().reset_index()
no_app = no_app[no_app['passage'] == df['passage'].max()].copy()
no_app['passage'] = df['passage'].max() + 1
app = | pd.concat([app, no_app]) | pandas.concat |
import hydra
from ncmw import community
from omegaconf import DictConfig, OmegaConf
import cobra
import logging
import socket
import time
import random
import numpy as np
import pandas as pd
from copy import deepcopy
import sys, os
import json, pickle
file_dir = os.path.dirname(os.path.dirname(__file__))
sys.path.append(file_dir)
from ncmw.utils.utils_io import (
get_models,
get_result_path,
SEPERATOR,
save_model,
get_model_paths,
)
from ncmw.visualization import (
plot_pairwise_growth_relation_per_weight,
plot_community_interaction,
plot_posterior_samples_for_observation,
plot_community_uptake_graph,
plot_species_interaction,
plot_community_summary,
plot_weight_growth_pairplot,
)
from ncmw.utils.utils_io import (
get_models,
get_result_path,
SEPERATOR,
save_model,
get_model_paths,
get_mediums,
)
from ncmw.community import (
BagOfReactionsModel,
ShuttleCommunityModel,
compute_fair_weights,
compute_community_summary,
compute_dominant_weights,
community_weight_posterior,
)
from ncmw.utils import (
get_models,
get_result_path,
check_for_substring_in_folder,
SEPERATOR,
)
@hydra.main(config_path="../../data/hydra", config_name="config.yaml")
def run_community_hydra(cfg: DictConfig) -> None:
run_community(cfg)
def create_community_folder_backbone(community_folder:str, project_folder:str) -> None:
"""Creates the backbone folder system used by the script
Args:
community_folder: Path to the community folder, here contents is placed
project_folder: Path to the parent directory
"""
try:
if not os.path.exists(project_folder):
os.mkdir(project_folder)
if not os.path.exists(community_folder):
os.mkdir(community_folder)
if not os.path.exists(community_folder + SEPERATOR + "community_models"):
os.mkdir(community_folder + SEPERATOR + "community_models")
if not os.path.exists(community_folder + SEPERATOR + "medium"):
os.mkdir(community_folder + SEPERATOR + "medium")
if not os.path.exists(community_folder + SEPERATOR + "weight_inference"):
os.mkdir(community_folder + SEPERATOR + "weight_inference")
if not os.path.exists(community_folder + SEPERATOR + "experiments_BagOfReactionsModel"):
os.mkdir(community_folder + SEPERATOR + "experiments_BagOfReactionsModel")
if not os.path.exists(community_folder + SEPERATOR + "experiments_ShuttleCommunityModel"):
os.mkdir(community_folder + SEPERATOR + "experiments_ShuttleCommunityModel")
except:
raise ValueError("Could not generate output directory, maybe we do not have permission's to do so?")
def load_old_configs(community_folder:str, cfg:str) -> DictConfig:
"""This will load the old config file to avoid recomputation.
Args:
community_folder: Folder in which the community results are placed.
cfg: Current config file to dump (is after this run the new old_config).
Returns:
DictConfig: Old configurations.
"""
if os.path.exists(community_folder + SEPERATOR + ".configs"):
with open(community_folder + SEPERATOR + ".configs", "rb") as f:
old_cfg = pickle.load(f)
else:
old_cfg = cfg
with open(community_folder + SEPERATOR + ".configs", "wb") as f:
pickle.dump(cfg, f)
return old_cfg
def set_solver_disable_functionalities_if_needed(cfg:DictConfig, log:logging.Logger) -> DictConfig:
"""This will set the solver to cplex, the default we recommend for this task.
Otherwise it will disable some functionality, which we encountered to be not
supported/hard for other solvers
Args:
cfg: Config file
log: Logger to return logs.
Returns:
DictConfig: Updated configs
"""
cobra_config = cobra.Configuration()
try:
cobra_config.solver = "cplex"
except Exception as e:
log.warn(f"We recommend cplex as solver, but it seems to be not installed on your system. We disable, cooperative tradeoff and community fva for other solvers. The error was {e}")
cfg.community.compute_community_fva = False
cfg.community.cooperative_tradeoff = False
return cfg
def generate_community_models(models, cfg, old_cfg, log, community_path):
log.info("Generating community models")
community_models = []
if cfg.community.bag_of_reactions_model:
path = (
community_path
+ SEPERATOR
+ "community_models"
+ SEPERATOR
+ "BagOfReactionsModel.pkl"
)
# Either loading or reinitializing the community model
if (
check_for_substring_in_folder(
community_path + SEPERATOR + "community_models", "BagOfReactionsModel.pkl"
)
and old_cfg.community.models_folder == cfg.community.models_folder
):
log.info("Loading BagOfReactions community model")
m = BagOfReactionsModel.load(path)
ids = [mod.id for mod in m.models]
correct = True
for model in models:
correct = correct and (model.id in ids)
if correct:
community_models += [m]
else:
community_models += [BagOfReactionsModel(models)]
else:
log.info("Building BagOfReactions Community model")
community_models += [BagOfReactionsModel(models)]
if cfg.community.shuttle_reactions_model:
path = (
community_path
+ SEPERATOR
+ "community_models"
+ SEPERATOR
+ "ShuttleCommunityModel.pkl"
)
kwargs = cfg.community.shuttle_reaction_params
if not isinstance(kwargs["shared_reactions"], list):
kwargs["shared_reactions"] = None
old_cfg.community.shuttle_reaction_params["shared_reactions"] = None
if (
check_for_substring_in_folder(
community_path + SEPERATOR + "community_models", "ShuttleCommunityModel.pkl"
)
and old_cfg.community.models_folder == cfg.community.models_folder
and all(
[
old_cfg.community.shuttle_reaction_params[key]
== cfg.community.shuttle_reaction_params[key]
for key in cfg.community.shuttle_reaction_params
]
)
):
log.info("Loading shuttle reactions model")
m = ShuttleCommunityModel.load(path)
ids = [mod.id for mod in models]
correct = True
for model in models:
correct = correct and (model.id in ids)
if correct:
community_models += [m]
else:
community_models += [ShuttleCommunityModel(models, **kwargs)]
else:
log.info("Building shuttle reactions model")
community_models += [ShuttleCommunityModel(models, **kwargs)]
log.info(f"Set correct weights: {cfg.community.main_weights}")
for m in community_models:
if cfg.community.main_weights == "ones":
weights = np.ones(len(m.models))
elif cfg.community.main_weights == "fair":
weights = compute_fair_weights(m)
else:
weights = cfg.community.main_weights
assert len(weights) == len(
m.models
), "The custom main weights must be a iterable of floats for each member of the community!"
m.weights = weights
return community_models
def reset(old_community_model, PATH):
new_model = type(old_community_model).load(PATH + SEPERATOR + "community_models" + SEPERATOR + str(type(old_community_model).__name__) + ".pkl")
new_model.medium = old_community_model.medium
new_model.weights = old_community_model.weights
return new_model
def generate_medias(community_model, cfg:DictConfig, log, community_path, result_path):
all_medias = {}
default = set(get_mediums("medium")[cfg.setup.medium].keys())
if cfg.community.medium["default"]:
log.info("Computing default for community")
# Also save default medium
medium = community_model.medium
if cfg.community.medium_strict_subset_of_default:
medium = dict([(k,v) for k,v in medium.items() if k in default])
all_medias["default"] = medium
path = community_path + SEPERATOR + "medium" + SEPERATOR + "DEFAULT" + ".json"
with open(path, "w+") as f:
json.dump(community_model.medium, f)
log.info(f"Saving default: {path}")
if cfg.community.medium["compm"] or cfg.community.medium["coopm"]:
log.info("Computing COMPM for community")
medium_prefix = result_path + SEPERATOR + "analysis" + SEPERATOR
mediums = get_mediums("medium", medium_prefix)
COMPM = dict()
for medium in mediums.values():
for key, val in medium.items():
COMPM[key] = float(abs(val))
all_medias["compm"] = COMPM
if cfg.community.medium_strict_subset_of_default:
COMPM = dict([(k,v) for k,v in COMPM.items() if k in default])
if cfg.community.medium["compm"]:
path = community_path + SEPERATOR + "medium" + SEPERATOR + "COMPM" + ".json"
with open(path, "w+") as f:
json.dump(COMPM, f)
log.info(f"Saving COMPMs: {path}")
return all_medias
def generate_coopm_medium(community_model, all_medias, cfg, log:logging.Logger, community_path):
log.info("Computing COOPM medium")
# COOPM computed using COMPM as base medium!
community_model.medium = all_medias["compm"]
mbr = community_model.slim_optimize()
if cfg.community.coopm_params["enforce_survival"] > 0:
for i in range(len(community_model.models)):
if community_model.single_optimize(i) == 0:
cfg.community.coopm_params["enforce_survival"] = 0
log.warning(
"We set enforce survival = 0, as not all models within the community can grow!"
)
log.info(f"Growth on COMPM: {mbr}")
coopm = community_model.compute_COOPM(mbr, **cfg.community.coopm_params)
all_medias["coopm"] = coopm
path = community_path + SEPERATOR + "medium" + SEPERATOR + f"{type(community_model).__name__}_COOPM" + ".json"
with open(path, "w+") as f:
json.dump(coopm, f)
log.info(f"Saving COOPMS: {path}")
return coopm
def flux_analysis(community_model, models, medium_name, cfg, log, PATH, path_to_save):
log.info(f"Community FBA/FVA results on medium: {medium_name}")
df_growth_summary = | pd.DataFrame() | pandas.DataFrame |
#!/usr/bin/env python
from . import net_feature_extract, snp_feature_extract, trace_feature_extract, utils
import podspy.log as logpkg
import podspy.petrinet as petripkg
import os, sys
import pandas as pd
import numpy as np
__all__ = [
'extract_feature_df'
]
@utils.timeit(on=True, verbose=False)
def extract_net_feature_ss(net):
net_feature_dict = net_feature_extract.extract_features(net)
net_feature_ss = pd.Series(net_feature_dict)
col_order = [
# net_feature_extract.N_TRAN,
# net_feature_extract.N_INV_TRAN,
# net_feature_extract.N_DUP_TRAN,
# net_feature_extract.N_UNIQ_TRAN,
# net_feature_extract.INV_TRAN_IN_DEG_MEAN,
# net_feature_extract.INV_TRAN_IN_DEG_STD,
# net_feature_extract.INV_TRAN_OUT_DEG_MEAN,
# net_feature_extract.INV_TRAN_OUT_DEG_STD,
# net_feature_extract.UNIQ_TRAN_IN_DEG_MEAN,
# net_feature_extract.UNIQ_TRAN_IN_DEG_STD,
# net_feature_extract.UNIQ_TRAN_OUT_DEG_MEAN,
# net_feature_extract.UNIQ_TRAN_OUT_DEG_STD,
# net_feature_extract.DUP_TRAN_IN_DEG_MEAN,
# net_feature_extract.DUP_TRAN_IN_DEG_STD,
# net_feature_extract.DUP_TRAN_OUT_DEG_MEAN,
# net_feature_extract.DUP_TRAN_OUT_DEG_STD,
# net_feature_extract.PLACE_IN_DEG_MEAN,
# net_feature_extract.PLACE_IN_DEG_STD,
# net_feature_extract.PLACE_OUT_DEG_MEAN,
# net_feature_extract.PLACE_OUT_DEG_STD,
# net_feature_extract.N_PLACE,
# net_feature_extract.N_ARC,
# net_feature_extract.N_AND_SPLIT,
# net_feature_extract.N_XOR_SPLIT,
# net_feature_extract.N_BICONNECTED_COMPONENT
net_feature_extract.PLACE_IN_DEG_MIN,
net_feature_extract.PLACE_IN_DEG_MAX,
net_feature_extract.PLACE_OUT_DEG_MIN,
net_feature_extract.PLACE_OUT_DEG_MAX,
net_feature_extract.INV_TRAN_IN_DEG_MIN,
net_feature_extract.INV_TRAN_IN_DEG_MAX,
net_feature_extract.INV_TRAN_OUT_DEG_MIN,
net_feature_extract.INV_TRAN_OUT_DEG_MAX,
net_feature_extract.DUP_TRAN_IN_DEG_MIN,
net_feature_extract.DUP_TRAN_IN_DEG_MAX,
net_feature_extract.DUP_TRAN_OUT_DEG_MIN,
net_feature_extract.DUP_TRAN_OUT_DEG_MAX,
net_feature_extract.UNIQ_TRAN_IN_DEG_MIN,
net_feature_extract.UNIQ_TRAN_IN_DEG_MAX,
net_feature_extract.UNIQ_TRAN_OUT_DEG_MIN,
net_feature_extract.UNIQ_TRAN_OUT_DEG_MAX,
net_feature_extract.N_PLACE_ONE_IN_DEG,
net_feature_extract.N_PLACE_ONE_OUT_DEG,
net_feature_extract.N_PLACE_TWO_IN_DEG,
net_feature_extract.N_PLACE_TWO_OUT_DEG,
net_feature_extract.N_PLACE_THREE_IN_DEG,
net_feature_extract.N_PLACE_THREE_OUT_DEG,
net_feature_extract.N_PLACE_MORE_THAN_FIVE_IN_DEG,
net_feature_extract.N_PLACE_MORE_THAN_FIVE_OUT_DEG,
net_feature_extract.N_INV_TRAN_ONE_IN_DEG,
net_feature_extract.N_INV_TRAN_ONE_OUT_DEG,
net_feature_extract.N_INV_TRAN_TWO_IN_DEG,
net_feature_extract.N_INV_TRAN_TWO_OUT_DEG,
net_feature_extract.N_INV_TRAN_THREE_IN_DEG,
net_feature_extract.N_INV_TRAN_THREE_OUT_DEG,
net_feature_extract.N_INV_TRAN_MORE_THAN_FIVE_IN_DEG,
net_feature_extract.N_INV_TRAN_MORE_THAN_FIVE_OUT_DEG,
net_feature_extract.N_DUP_TRAN_ONE_IN_DEG,
net_feature_extract.N_DUP_TRAN_ONE_OUT_DEG,
net_feature_extract.N_DUP_TRAN_TWO_IN_DEG,
net_feature_extract.N_DUP_TRAN_TWO_OUT_DEG,
net_feature_extract.N_DUP_TRAN_THREE_IN_DEG,
net_feature_extract.N_DUP_TRAN_THREE_OUT_DEG,
net_feature_extract.N_DUP_TRAN_MORE_THAN_FIVE_IN_DEG,
net_feature_extract.N_DUP_TRAN_MORE_THAN_FIVE_OUT_DEG,
net_feature_extract.N_UNIQ_TRAN_ONE_IN_DEG,
net_feature_extract.N_UNIQ_TRAN_ONE_OUT_DEG,
net_feature_extract.N_UNIQ_TRAN_TWO_IN_DEG,
net_feature_extract.N_UNIQ_TRAN_TWO_OUT_DEG,
net_feature_extract.N_UNIQ_TRAN_THREE_IN_DEG,
net_feature_extract.N_UNIQ_TRAN_THREE_OUT_DEG,
net_feature_extract.N_UNIQ_TRAN_MORE_THAN_FIVE_IN_DEG,
net_feature_extract.N_UNIQ_TRAN_MORE_THAN_FIVE_OUT_DEG
]
net_feature_ss = net_feature_ss[col_order]
return net_feature_ss
def extract_decomposition_feature_ss(decomposition):
feature_dict = net_feature_extract.extract_features_from_decomposition(decomposition)
feature_ss = | pd.Series(feature_dict) | pandas.Series |
"""Integer optimization of livestock and water services."""
import os
import sys
import shutil
from osgeo import gdal
import re
import pandas
import numpy as np
import pygeoprocessing.geoprocessing
import marginal_value as mv
def integer_optim_Peru(objective_list, suf):
"""Calculate optimal intervention portfolio for a set of objective weights.
Parameters:
objective_list (list): list of objective objects containing info about
each objective used to construct the optimization problem including
objective weight, target, and whether it should be maximized or not
suf (string): results suffix that will be appended to the filename of
solution
Side effects:
creates or modifies a csv file containing the solution, the optimal
intervention set
creates or modifies a csv file containing scores, objective scores for
the optimal intervention set
Returns:
solution_filename, path to csv file where solution was saved
"""
intervention_list = [
'camelid_high', 'camelid_high_rot', 'camelid_low', 'camelid_low_rot',
'cow_high', 'cow_high_rot', 'cow_low', 'cow_lot_rot',
'sheep_high', 'sheep_high_rot', 'sheep_low', 'sheep_low_rot']
pdict = {
u'outerdir': os.path.join(
_DATA_INPUT_DIR, 'animal_weights_literature_default_beta'),
u'rau_shp': os.path.join(_DATA_INPUT_DIR, 'canete_basin.shp'),
u'lulc': os.path.join(_DATA_INPUT_DIR, 'Final_cobertura_Canete.tif')}
intermediate_dir = os.path.join(pdict[u'outerdir'], 'intermediate')
if not os.path.exists(intermediate_dir):
os.makedirs(intermediate_dir)
pdict[u'intermediate'] = intermediate_dir
output_dir = os.path.join(pdict[u'outerdir'], 'output')
if not os.path.exists(output_dir):
os.makedirs(output_dir)
pdict[u'output'] = output_dir
rau = 0
csv_folder = os.path.join(pdict['outerdir'], 'marginal_value_csvs')
tables_folder = 'integer_optimizer_data'
def generate_ll_input_data():
tables_list = mv.margv_tables_from_csv(
pdict, objective_list, csv_folder, tables_folder)
# move marginal value tables just generated
for obj in objective_list:
int_folder = os.path.join(intermediate_dir, 'rau_%s' % obj.name)
if not os.path.exists(int_folder):
os.makedirs(int_folder)
copyfrom = os.path.join(
intermediate_dir, tables_folder, '%s_rau0.npy' % obj.name)
copyto = os.path.join(int_folder, '%s_rau0.npy' % obj.name)
shutil.copyfile(copyfrom, copyto)
copyto = os.path.join(int_folder, 'rau0.npy')
shutil.copyfile(copyfrom, copyto)
# normalize values
for objective in objective_list:
if objective.name == 'cost' or objective.name == 'Cost':
continue
folder = os.path.join(
pdict[u'intermediate'], 'rau_' + objective.name)
mv.normalize_values(folder, objective.maximize)
# rename normalized arrays
for obj in objective_list:
copyfrom = os.path.join(
pdict[u'intermediate'], 'rau_%s' % obj.name, 'norm',
'%s_rau0.npy' % obj.name)
copyto = os.path.join(
pdict[u'intermediate'], 'rau_%s' % obj.name, 'norm',
'rau0.npy')
shutil.move(copyfrom, copyto)
generate_ll_input_data()
ll_problem = {
'weights': {},
'targets': {},
'targettypes': {}}
for objective in objective_list:
ll_problem['weights'][objective.name] = objective.weight
if objective.l_target is not None:
ll_problem['targets'][objective.name] = objective.l_target
if objective.target_type is not None:
ll_problem['targettypes'][objective.name] = objective.target_type
ll_data = {'factornames': []}
for objective in objective_list:
ll_data['factornames'].append(objective.name)
if objective.name == 'cost' or objective.name == 'Cost':
rau_dir = os.path.join(
pdict[u'intermediate'], 'rau_' + objective.name)
else:
rau_dir = os.path.join(
pdict[u'intermediate'], 'rau_' + objective.name, 'norm')
file = os.path.join(rau_dir, 'rau' + str(rau) + '.npy')
if not os.path.isfile(file):
raise Exception("file %s not found" % file)
ll_data[objective.name] = np.load(file)
# get un-normalized objective data
ll_marg_data = ll_data.copy()
for objective in objective_list:
rau_dir = os.path.join(
pdict[u'intermediate'], 'rau_' + objective.name)
file = os.path.join(rau_dir, 'rau' + str(rau) + '.npy')
if not os.path.isfile(file):
raise Exception("file %s not found" % file)
ll_marg_data[objective.name] = np.load(file)
solution, scores = mv.integer_optimization(
ll_data, ll_problem, rau, marg_data=ll_marg_data,
tiebreaker_intervention=0)
solution_filename = os.path.join(
pdict['outerdir'], 'output', 'solution%s.csv' % suf)
scores_filename = os.path.join(
pdict['outerdir'], 'output', 'scores%s.csv' % suf)
solution_df = pandas.DataFrame({'solution': solution})
solution_df.to_csv(solution_filename)
scores_df = pandas.DataFrame(scores, index=[0])
scores_df.to_csv(scores_filename)
return solution_filename
def translate_soln_to_lulc(solution_table, out_name):
"""Generate landcover raster from one optimal solution.
Parameters:
solution_table (string): path to csv table containing optimal
intervention portfolio according to one set of objective weights
out_name (string): file location where landcover raster should be
saved
Side effects:
creates or modifies a geotiff at the location `out_name`
Returns:
None
"""
hru_lulc_table = os.path.join(_DATA_INPUT_DIR, 'hru_definition_table.csv')
HRU_raster = os.path.join(_DATA_INPUT_DIR, 'HRU_all.tif')
HRU_codes = os.path.join(_DATA_INPUT_DIR, 'HRU_codes_11.8.16.csv')
sol_df = pandas.read_csv(solution_table)
HRU_df = pandas.read_csv(HRU_codes)
sol_joined = pandas.concat([sol_df, HRU_df], axis=1)
out_datatype = gdal.GDT_Int32
source_dataset = gdal.Open(HRU_raster)
band = source_dataset.GetRasterBand(1)
out_nodata = band.GetNoDataValue()
lulc_df = pandas.read_csv(hru_lulc_table)
merged_df = pandas.merge(sol_joined, lulc_df, on='HRU', how='outer')
merged_df['soln_int'] = merged_df['solution'].astype(float)
merged_df['sb_lu'] = merged_df['sb_lu'].astype(float)
merged_df.loc[
merged_df['solution'].notnull(), 'new_lulc'] = merged_df[
'sb_lu'] * 100 + merged_df['soln_int']
merged_df.loc[merged_df['solution'].isnull(), 'new_lulc'] = merged_df[
'sb_lu']
value_map = {row[3]: row[9] for row in merged_df.itertuples()}
pygeoprocessing.geoprocessing.reclassify_dataset_uri(
HRU_raster, value_map, out_name, out_datatype, out_nodata)
band = None
del source_dataset
def translate_solution(solution_csv, HRU_codes, HRU_raster, raster_out_uri):
"""Create raster showing optimal intervention for each HRU.
Parameters:
solution_csv (string): path to csv file containing the optimal
intervention portfolio according to one set of objective weights
HRU_codes (string): path to csv file containing HRU codes in a single
column. This file can be created by copying the 'zone' column from
one of the marginal value rasters (e.g.,
livestock_mv_by_HRU_9.29.16.csv).
HRU_raster (string): path to geotiff containing hydrologic response
units (HRUs) indexed to the integer codes in `HRU_codes`
raster_out_uri (string): path to location on disk where optimal
intervention geotiff should be saved
Side effects:
Creates or modifies the geotiff located at `raster_out_uri`
Returns:
None
"""
HRU_list = pygeoprocessing.geoprocessing.unique_raster_values_uri(
HRU_raster)
sol_df = pandas.read_csv(solution_csv)
HRU_df = pandas.read_csv(HRU_codes)
assert len(set(HRU_list) - set(HRU_df.HRU)) == 0, """Error: HRU raster does
not match HRU codes"""
sol_joined = pandas.concat([sol_df, HRU_df], axis=1)
out_datatype = 3
source_dataset = gdal.Open(HRU_raster)
band = source_dataset.GetRasterBand(1)
out_nodata = band.GetNoDataValue()
value_map = {row[3]: row[2] for row in sol_joined.itertuples()}
pygeoprocessing.geoprocessing.reclassify_dataset_uri(
HRU_raster, value_map, raster_out_uri, out_datatype, out_nodata)
band = None
del source_dataset
def integer_optim_wrapper():
"""Run integer optimization at series of objective weight combinations.
This function calls `integer_optim_Peru`, `translate_solution`, and
`translate_soln_to_lulc` to calculate optimal interventions for a series of
objective weights and generate tables and maps from that solution.
Side effects:
creates or modifies files located at hard-coded locations on disk
Returns:
None
"""
weight_range = [0, 0.1, 0.3, 0.5, 0.7, 0.9, 1]
for livestock_weight in weight_range:
for sdr_weight in weight_range:
for swy_weight in weight_range:
if (livestock_weight == sdr_weight and
sdr_weight == swy_weight):
if swy_weight < 1: # only run once with equal weights
continue
if (livestock_weight == 0 and sdr_weight == 0 and
swy_weight == 0):
continue
sed_obj = mv.Objective(
'sdr', sdr_weight, None, None, None, maximize=False)
swy_obj = mv.Objective(
'swy', swy_weight, None, None, None, maximize=True)
livestock_obj = mv.Objective(
'livestock', livestock_weight, None, None, None,
maximize=True)
objective_list = [sed_obj, swy_obj, livestock_obj]
suf = 'livestock_{}_sdr_{}_swy_{}'.format(
livestock_obj.weight, sed_obj.weight, swy_obj.weight)
raster_out_uri = os.path.join(
_DATA_INPUT_DIR, 'animal_weights_literature_default_beta',
'output', 'solution_map{}'.format(suf))
if not os.path.exists(raster_out_uri):
solution_csv = integer_optim_Peru(objective_list, suf)
HRU_codes = os.path.join(
_DATA_INPUT_DIR, 'HRU_codes_11.8.16.csv')
HRU_raster = os.path.join(
_DATA_INPUT_DIR, 'HRU_priority_FESC_RYEG.tif')
translate_solution(
solution_csv, HRU_codes, HRU_raster, raster_out_uri)
lulc_out_name = os.path.join(
_DATA_INPUT_DIR,
'animal_weights_literature_default_beta',
'output', 'solution_lulc{}.tif'.format(suf))
translate_soln_to_lulc(solution_csv, lulc_out_name)
def collate_scores(output_folder, save_as):
"""Collect scores into a file for plotting frontiers.
Parameters:
output_folder (string): path to local file folder containing a series
of csv tables, one for each objective weight combination,
indicating objective scores for the optimal solution
save_as (string): path to location where summary of objective scores
across objective weights should be saved
Side effects:
creates or modifies the csv table indicated by the path `save_as`
Returns:
None
"""
scores_files = [
f for f in os.listdir(output_folder) if f.startswith('scores')]
f = scores_files[0]
sol_df = pandas.read_csv(os.path.join(output_folder, f))
objectives = sol_df.columns.values.tolist()
objectives.remove('objective')
objectives.remove('Unnamed: 0')
sum_dict = {}
for obj in objectives:
sum_dict['{}_weight'.format(obj)] = []
sum_dict['{}_score'.format(obj)] = []
for f in scores_files:
sol_df = pandas.read_csv(os.path.join(output_folder, f))
for obj in objectives:
score = sol_df.get_value(0, obj)
try:
pattern = '{}_(.+?)_'.format(obj)
weight = re.search(pattern, f).group(1)
except IndexError:
pattern = '{}_(.+?).csv'.format(obj)
weight = re.search(pattern, f).group(1)
sum_dict['{}_weight'.format(obj)].append(weight)
sum_dict['{}_score'.format(obj)].append(score)
sum_df = pandas.DataFrame(sum_dict)
sum_df.to_csv(save_as)
def collate_solutions(output_folder, objective_list):
"""Collect solutions from several portfolios.
Parameters:
output_folder (string): path to directory on disk that contains
solution summaries
objective_list (list): list of strings identifying the order of
objectives
Side effects:
creates or modifies the following files in `output_folder`:
'solution_summary.csv'
'solution_index.csv'
Returns:
None
"""
solutions_files = [
f for f in os.listdir(output_folder) if f.startswith('solution') and
f.endswith('.csv')]
df_list = []
weight_dict = {obj: [] for obj in objective_list}
weight_dict['soln_index'] = []
for idx in xrange(len(solutions_files)):
f = solutions_files[idx]
weight_dict['soln_index'].append(idx)
for obj in objective_list:
try:
pattern = '{}_(.+?)_'.format(obj)
weight = re.search(pattern, f).group(1)
except IndexError:
pattern = '{}_(.+?).csv'.format(obj)
weight = re.search(pattern, f).group(1)
weight_dict[obj].append(weight)
df = pandas.read_csv(os.path.join(output_folder, f))
del df['Unnamed: 0']
df.columns = [idx]
df_list.append(df)
result_df = pandas.concat(df_list, axis=1)
weight_df = pandas.DataFrame(weight_dict)
result_df.to_csv(os.path.join(output_folder, "solution_summary.csv"),
index=False)
weight_df.to_csv(os.path.join(output_folder, "solution_index.csv"),
index=False)
def solution_agreement(HRU_codes, solution_summary, save_as):
"""Calculate agreement metrics from a set of portfolios.
Parameters:
HRU_codes (string): path to csv file containing HRU codes in a single
column of integers
solution_summary (string): path to csv file containing summary of
solutions among objective weight combinations
save_as (string): path to location on disk where agreement metrics
should be saved
Side effects:
creates or modifies the csv file indicated by `save_as`
Returns:
None
"""
sol_df = pandas.read_csv(solution_summary)
stat_df = pandas.read_csv(HRU_codes)
# proportion of runs where each HRU was selected for intervention
stat_df['prop_selected'] = (
sol_df.astype(bool).sum(axis=1) / sol_df.shape[1])
# most often chosen intervention: mode
stat_df['mode'] = stats.mode(sol_df, axis=1)[0]
stat_df.set_index("HRU")
stat_df.to_csv(save_as, index=False)
def create_agreement_rasters(agreement_summary, HRU_raster, raster_out_dir):
"""Make rasters of agreement metrics.
Parameters:
agreement_summary (string): path to csv containing portfolio agreement
metrics
HRU_raster (string): path to geotiff containing HRUs
raster_out_dir (string): path to location on disk where agreement
rasters should be saved
Side effects:
creates or modifies the following files in `raster_out_dir`:
'proportion_selected.tif'
'"most_often_chosen.tif'
Returns:
None
"""
HRU_list = pygeoprocessing.geoprocessing.unique_raster_values_uri(
HRU_raster)
agreement_df = | pandas.read_csv(agreement_summary) | pandas.read_csv |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
# @Date : Feb-03-20 23:44
# @Author : <NAME> (<EMAIL>)
# @Link : http://example.org
import time
import os
import json
import random
import pickle
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping, LearningRateScheduler, ModelCheckpoint, ReduceLROnPlateau, CSVLogger
from tensorflow.keras.preprocessing.image import ImageDataGenerator, load_img
from tensorflow.keras.metrics import AUC, BinaryAccuracy
from resnet.resnet import model_depth, resnet_v2, lr_schedule
# Parameters we care
exper_name = "ResNet56v2_origin"
test_on_train = True
START_EPOCH = 150 # 已经完成的训练数
ALPHA = 0.99 # label 1 sample's weight
BATCH_SIZE = 32 # 16 for Mac, 64, 128 for server
IF_FAST_RUN = True # False
# Training parameters
TRAINING_EPOCHS = 150
TOTAL_TRAIN = 30000 * 0.8
TOTAL_VALIDATE = 30000 * 0.2
# constants
IF_DATA_AUGMENTATION = True
NUM_CLASSES = 2
IMAGE_WIDTH = IMAGE_HEIGHT = 224
IMAGE_SIZE = (IMAGE_WIDTH, IMAGE_HEIGHT)
IMAGE_CHANNELS = 1
INPUT_SHAPE = [IMAGE_WIDTH, IMAGE_HEIGHT, IMAGE_CHANNELS]
METRICS = [
BinaryAccuracy(name='accuracy'), # 整体的 accuracy
AUC(name='auc_good_0') # 以 good 为 positive 的 AUC
]
print("If in eager mode: ", tf.executing_eagerly())
print("Use tensorflow version 2.")
assert tf.__version__[0] == "2"
print("Load config ...")
with open('./config/config_win.json', 'r') as f:
CONFIG = json.load(f)
ROOT_PATH = CONFIG["ROOT_PATH"]
print(f"ROOT_PATH: {ROOT_PATH}")
ROOT_PATH = os.path.expanduser(ROOT_PATH)
print(f"ROOT_PATH: {ROOT_PATH}")
TRAIN_DATA_DIR = os.path.join(ROOT_PATH, CONFIG["TRAIN_DATA_DIR"])
print(f"TRAIN_DATA_DIR: {TRAIN_DATA_DIR}")
TEST_DATA_DIR = os.path.join(ROOT_PATH, CONFIG["TEST_DATA_DIR"])
print(f"TEST_DATA_DIR: {TEST_DATA_DIR}")
print("Prepare testing data...")
if test_on_train:
num_samples = num_train = 30000
label_names = os.listdir(TRAIN_DATA_DIR)
filenames, labels = [], []
for i, label in enumerate(label_names):
files = os.listdir(os.path.join(TRAIN_DATA_DIR, label))
for f in files:
filenames.append(label+"/"+f)
labels.append(i) # 0 or 1
table = np.asarray([filenames, labels])
table = table.T
columns = ["filename", "label"]
# test on train dataset
test_df = | pd.DataFrame(data=table, columns=columns) | pandas.DataFrame |
import logging
from unittest.mock import Mock
import pandas as pd
import pytest
from numpy import nan
from pdlog.logging import log_change_index
from pdlog.logging import log_fillna
from pdlog.logging import log_filter
from pdlog.logging import log_rename
from pdlog.logging import log_reshape
@pytest.fixture
def caplog(caplog):
caplog.set_level(logging.INFO)
return caplog
def _test_log_function(
log_fn, caplog, before_df, after_df, expected_level, expected_msg
):
fn_args = Mock()
fn_kwargs = Mock()
before_df.fn = Mock(return_value=after_df)
log_fn(before_df, "fn", fn_args, fn_kwargs)
before_df.fn.assert_called_once_with(fn_args, fn_kwargs)
assert len(caplog.records) == 1
record = caplog.records[0]
assert record.levelno == expected_level
assert record.message == expected_msg
@pytest.mark.parametrize(
("before", "after", "expected_level", "expected_msg"),
(
pytest.param([], [], logging.INFO, "fn: empty input dataframe", id="empty_df"),
pytest.param(
[0, 1, 2], [], logging.CRITICAL, "fn: dropped all rows", id="all_rows"
),
pytest.param(
{"x": [1, 2, 3], "y": [4, 5, 6], "z": [7, 8, 9]},
{"x": [1]},
logging.INFO,
"fn: dropped 2 columns (67%) and 2 rows (67%), 1 row remaining",
id="some_rows_and_cols",
),
pytest.param(
[0, 1, 2],
[0, 1],
logging.INFO,
"fn: dropped 1 row (33%), 2 rows remaining",
id="some_rows",
),
pytest.param(
{"x": [1, 2, 3], "y": [4, 5, 6], "z": [7, 8, 9]},
{"x": [1, 2, 3]},
logging.INFO,
"fn: dropped 2 columns (67%): ['y', 'z']",
id="some_cols",
),
pytest.param(
[0, 1, 2], [0, 1, 2], logging.INFO, "fn: dropped no rows", id="no_rows"
),
),
)
def test_log_filter(caplog, before, after, expected_level, expected_msg):
before_df = pd.DataFrame(before)
after_df = pd.DataFrame(after)
_test_log_function(
log_filter, caplog, before_df, after_df, expected_level, expected_msg
)
@pytest.mark.parametrize(
("before", "after", "error", "error_msg"),
[
pytest.param(
[],
[0, 1, 2],
AssertionError,
"function: fn added rows, it is not a valid filter operation",
id="add_rows",
),
pytest.param(
{"x": [1, 2, 3]},
{"x": [1, 2, 3], "y": [4, 5, 6], "z": [7, 8, 9]},
AssertionError,
"function: fn added columns, it is not a valid filter operation",
id="add_cols",
),
],
)
def test_log_filter_raises(before, after, error, error_msg):
before_df = pd.DataFrame(before)
after_df = pd.DataFrame(after)
before_df.fn = Mock(return_value=after_df)
with pytest.raises(error, match=error_msg):
log_filter(before_df, "fn")
@pytest.mark.parametrize(
("before", "after", "expected_level", "expected_msg"),
(
(
pd.RangeIndex(3),
pd.date_range("2020-01-01", "2020-01-03", name="date"),
logging.INFO,
(
"fn: set from 'None' (RangeIndex): [0, 1, 2] to 'date' "
"(DatetimeIndex): ['2020-01-01 00:00:00', "
"'2020-01-02 00:00:00', '2020-01-03 00:00:00']"
),
),
),
)
def test_log_change_index(caplog, before, after, expected_level, expected_msg):
before_df = pd.DataFrame(index=before)
after_df = pd.DataFrame(index=after)
_test_log_function(
log_change_index, caplog, before_df, after_df, expected_level, expected_msg
)
@pytest.mark.parametrize(
(
"before_index",
"before_columns",
"after_index",
"after_columns",
"expected_level",
"expected_msg",
),
(
pytest.param(
["foo", "bar", "baz"],
["foo", "bar", "baz"],
["foo_1", "bar", "baz"],
["foo", "bar_1", "baz_1"],
logging.INFO,
(
"fn: renamed 1 row and 2 columns. "
"rows: ['foo_1']. columns: ['bar_1', 'baz_1']"
),
id="rows_and_columns",
),
pytest.param(
["foo", "bar", "baz"],
[],
["foo", "bar_1", "baz_1"],
[],
logging.INFO,
"fn: renamed 2 rows: ['bar_1', 'baz_1']",
id="rows",
),
pytest.param(
[],
["foo", "bar", "baz"],
[],
["foo", "bar_1", "baz_1"],
logging.INFO,
"fn: renamed 2 columns: ['bar_1', 'baz_1']",
id="columns",
),
pytest.param(
["foo", "bar", "baz"],
["foo", "bar", "baz"],
["foo", "bar", "baz"],
["foo", "bar", "baz"],
logging.INFO,
"fn: renamed nothing",
id="nothing",
),
),
)
def test_log_rename(
caplog,
before_index,
after_index,
before_columns,
after_columns,
expected_level,
expected_msg,
):
before_df = pd.DataFrame(index=before_index, columns=before_columns)
after_df = | pd.DataFrame(index=after_index, columns=after_columns) | pandas.DataFrame |
import numpy as np
import pytest
from pandas._libs import join as _join
from pandas import Categorical, DataFrame, Index, merge
import pandas._testing as tm
class TestIndexer:
@pytest.mark.parametrize(
"dtype", ["int32", "int64", "float32", "float64", "object"]
)
def test_outer_join_indexer(self, dtype):
indexer = _join.outer_join_indexer
left = np.arange(3, dtype=dtype)
right = np.arange(2, 5, dtype=dtype)
empty = np.array([], dtype=dtype)
result, lindexer, rindexer = indexer(left, right)
assert isinstance(result, np.ndarray)
assert isinstance(lindexer, np.ndarray)
assert isinstance(rindexer, np.ndarray)
tm.assert_numpy_array_equal(result, np.arange(5, dtype=dtype))
exp = np.array([0, 1, 2, -1, -1], dtype=np.int64)
tm.assert_numpy_array_equal(lindexer, exp)
exp = np.array([-1, -1, 0, 1, 2], dtype=np.int64)
tm.assert_numpy_array_equal(rindexer, exp)
result, lindexer, rindexer = indexer(empty, right)
tm.assert_numpy_array_equal(result, right)
exp = np.array([-1, -1, -1], dtype=np.int64)
tm.assert_numpy_array_equal(lindexer, exp)
exp = np.array([0, 1, 2], dtype=np.int64)
tm.assert_numpy_array_equal(rindexer, exp)
result, lindexer, rindexer = indexer(left, empty)
tm.assert_numpy_array_equal(result, left)
exp = np.array([0, 1, 2], dtype=np.int64)
tm.assert_numpy_array_equal(lindexer, exp)
exp = np.array([-1, -1, -1], dtype=np.int64)
tm.assert_numpy_array_equal(rindexer, exp)
def test_left_join_indexer_unique():
a = np.array([1, 2, 3, 4, 5], dtype=np.int64)
b = np.array([2, 2, 3, 4, 4], dtype=np.int64)
result = _join.left_join_indexer_unique(b, a)
expected = np.array([1, 1, 2, 3, 3], dtype=np.int64)
tm.assert_numpy_array_equal(result, expected)
def test_left_outer_join_bug():
left = np.array(
[
0,
1,
0,
1,
1,
2,
3,
1,
0,
2,
1,
2,
0,
1,
1,
2,
3,
2,
3,
2,
1,
1,
3,
0,
3,
2,
3,
0,
0,
2,
3,
2,
0,
3,
1,
3,
0,
1,
3,
0,
0,
1,
0,
3,
1,
0,
1,
0,
1,
1,
0,
2,
2,
2,
2,
2,
0,
3,
1,
2,
0,
0,
3,
1,
3,
2,
2,
0,
1,
3,
0,
2,
3,
2,
3,
3,
2,
3,
3,
1,
3,
2,
0,
0,
3,
1,
1,
1,
0,
2,
3,
3,
1,
2,
0,
3,
1,
2,
0,
2,
],
dtype=np.int64,
)
right = np.array([3, 1], dtype=np.int64)
max_groups = 4
lidx, ridx = _join.left_outer_join(left, right, max_groups, sort=False)
exp_lidx = np.arange(len(left), dtype=np.int64)
exp_ridx = -np.ones(len(left), dtype=np.int64)
exp_ridx[left == 1] = 1
exp_ridx[left == 3] = 0
tm.assert_numpy_array_equal(lidx, exp_lidx)
tm.assert_numpy_array_equal(ridx, exp_ridx)
def test_inner_join_indexer():
a = np.array([1, 2, 3, 4, 5], dtype=np.int64)
b = np.array([0, 3, 5, 7, 9], dtype=np.int64)
index, ares, bres = _join.inner_join_indexer(a, b)
index_exp = np.array([3, 5], dtype=np.int64)
tm.assert_almost_equal(index, index_exp)
aexp = np.array([2, 4], dtype=np.int64)
bexp = np.array([1, 2], dtype=np.int64)
tm.assert_almost_equal(ares, aexp)
tm.assert_almost_equal(bres, bexp)
a = np.array([5], dtype=np.int64)
b = np.array([5], dtype=np.int64)
index, ares, bres = _join.inner_join_indexer(a, b)
tm.assert_numpy_array_equal(index, np.array([5], dtype=np.int64))
tm.assert_numpy_array_equal(ares, np.array([0], dtype=np.int64))
tm.assert_numpy_array_equal(bres, np.array([0], dtype=np.int64))
def test_outer_join_indexer():
a = np.array([1, 2, 3, 4, 5], dtype=np.int64)
b = np.array([0, 3, 5, 7, 9], dtype=np.int64)
index, ares, bres = _join.outer_join_indexer(a, b)
index_exp = np.array([0, 1, 2, 3, 4, 5, 7, 9], dtype=np.int64)
tm.assert_almost_equal(index, index_exp)
aexp = np.array([-1, 0, 1, 2, 3, 4, -1, -1], dtype=np.int64)
bexp = np.array([0, -1, -1, 1, -1, 2, 3, 4], dtype=np.int64)
tm.assert_almost_equal(ares, aexp)
tm.assert_almost_equal(bres, bexp)
a = np.array([5], dtype=np.int64)
b = np.array([5], dtype=np.int64)
index, ares, bres = _join.outer_join_indexer(a, b)
tm.assert_numpy_array_equal(index, np.array([5], dtype=np.int64))
tm.assert_numpy_array_equal(ares, np.array([0], dtype=np.int64))
tm.assert_numpy_array_equal(bres, np.array([0], dtype=np.int64))
def test_left_join_indexer():
a = np.array([1, 2, 3, 4, 5], dtype=np.int64)
b = np.array([0, 3, 5, 7, 9], dtype=np.int64)
index, ares, bres = _join.left_join_indexer(a, b)
tm.assert_almost_equal(index, a)
aexp = np.array([0, 1, 2, 3, 4], dtype=np.int64)
bexp = np.array([-1, -1, 1, -1, 2], dtype=np.int64)
tm.assert_almost_equal(ares, aexp)
tm.assert_almost_equal(bres, bexp)
a = np.array([5], dtype=np.int64)
b = np.array([5], dtype=np.int64)
index, ares, bres = _join.left_join_indexer(a, b)
tm.assert_numpy_array_equal(index, np.array([5], dtype=np.int64))
tm.assert_numpy_array_equal(ares, np.array([0], dtype=np.int64))
tm.assert_numpy_array_equal(bres, np.array([0], dtype=np.int64))
def test_left_join_indexer2():
idx = Index([1, 1, 2, 5])
idx2 = Index([1, 2, 5, 7, 9])
res, lidx, ridx = _join.left_join_indexer(idx2.values, idx.values)
exp_res = np.array([1, 1, 2, 5, 7, 9], dtype=np.int64)
tm.assert_almost_equal(res, exp_res)
exp_lidx = np.array([0, 0, 1, 2, 3, 4], dtype=np.int64)
tm.assert_almost_equal(lidx, exp_lidx)
exp_ridx = np.array([0, 1, 2, 3, -1, -1], dtype=np.int64)
tm.assert_almost_equal(ridx, exp_ridx)
def test_outer_join_indexer2():
idx = Index([1, 1, 2, 5])
idx2 = Index([1, 2, 5, 7, 9])
res, lidx, ridx = _join.outer_join_indexer(idx2.values, idx.values)
exp_res = np.array([1, 1, 2, 5, 7, 9], dtype=np.int64)
tm.assert_almost_equal(res, exp_res)
exp_lidx = np.array([0, 0, 1, 2, 3, 4], dtype=np.int64)
tm.assert_almost_equal(lidx, exp_lidx)
exp_ridx = np.array([0, 1, 2, 3, -1, -1], dtype=np.int64)
tm.assert_almost_equal(ridx, exp_ridx)
def test_inner_join_indexer2():
idx = Index([1, 1, 2, 5])
idx2 = Index([1, 2, 5, 7, 9])
res, lidx, ridx = _join.inner_join_indexer(idx2.values, idx.values)
exp_res = np.array([1, 1, 2, 5], dtype=np.int64)
tm.assert_almost_equal(res, exp_res)
exp_lidx = np.array([0, 0, 1, 2], dtype=np.int64)
tm.assert_almost_equal(lidx, exp_lidx)
exp_ridx = np.array([0, 1, 2, 3], dtype=np.int64)
tm.assert_almost_equal(ridx, exp_ridx)
def test_merge_join_categorical_multiindex():
# From issue 16627
a = {
"Cat1": | Categorical(["a", "b", "a", "c", "a", "b"], ["a", "b", "c"]) | pandas.Categorical |
# Import required modules
import requests
import pandas as pd
import json
import subprocess
from tqdm import tqdm
import re
# Set pandas to show full rows and columns
pd.set_option('display.max_rows', None)
pd.set_option('display.max_columns', None)
pd.set_option('display.width', None)
pd.set_option('display.max_colwidth', None)
'''
This is API Query Function
'''
# Main function starts here
def query(cname="cname", apikey="apikey", apiquery="apiquery"):
'''
-- cname --> Cloud Instance Name <cname.infocyte.com> ('cname' is without .infocyte.com)
-- apikey --> APIKEY or the API Token
-- apiquery --> API GET Method
** Set above args as pre-defined variables (Can be used multiple times) or call them on the fly (Single use).
icdata = ic.query(cname, apikey, apiquery)
variable 'icdata' --> PandasDataframe. Can now be used with all options available from pandas. Refer README and Wiki for more details. https://pandas.pydata.org/docs/
Note: 'query' function loops until it reaches the last page on API explorer. Larger the data, more time it takes. However each loop will pull 1K entries (rows) and progress details are displayed while quering the data.
'''
tqdm.pandas()
global icpd, icd
icd = requests.get("https://"+cname+".infocyte.com/api/" +
apiquery+"?access_token="+apikey + "&count=True")
if "There is no method to handle GET" in icd.content.decode("utf-8"):
print("API Endpoint not found, suffix \"/explorer/#/\" at the end of URL to find the correct end point")
elif icd.reason == "Not Found":
print("Please check the CNAME used is correct")
elif icd.reason == "Unauthorized":
print("Please check the APIKey / Token has the permission to access the instance")
elif icd.reason != "OK":
print("Something went wrong and unable to find the reason")
else:
iccount = (str(icd.headers.get("X-Total-Count"))[:-3])
if (len(iccount) == 0):
loopic = icd
for x in tqdm(range(1), desc="Loading " + apiquery, ncols=100, unit='Loop(s)', bar_format='{l_bar}{bar} | {n_fmt}/{total_fmt} {unit}', colour='GREEN'):
icdata = json.loads(loopic.text)
icdb = | pd.DataFrame(icdata) | pandas.DataFrame |
"""
This module implements several methods for calculating and outputting solutions of the unionfind_cluster_editing() algorithm.
It contains some methods to print solutions
and, more importantly, methods to merge solutions into one better solution.
There are 3 main algorithms: merge, repair and undo.
Two repair algorithms with differing complexity are implemented.
"""
from union_find import *
from math import log
import sys
import numpy as np
from numba import njit, jit
from numpy import random as rand
from model_sqrt import *
from numba.typed import Dict
import pandas as pd
def print_result(output_path, name, date):
file = open(output_path + name, mode="a")
file.write(str(date))
file.close()
def print_zhk(output_path, merged, sizes):
file = open(output_path + "zhk_sizes.txt", mode="a")
for i in range(len(sizes)):
if merged[i] == i:
file.write(str(i) + " " + str(sizes[i]) + "\n")
file.close()
def print_solution_costs(solution_costs, output_path):
"""
This function outputs all sorted solution costs to a ifle named "..._solution_costs.txt".
"""
sorted_costs = np.sort(solution_costs)
print_to = output_path + "solutions_v4.txt"
with open(print_to, mode="a") as file:
for cost in sorted_costs:
file.write(str(cost))
file.write("\n")
def all_solutions(solution_costs, parents, filename, missing_weight, n):
"""
This function outputs all solutions, sorted by their costs, to a ifle named "all_solutions.txt".
"""
cost_sorted_i = np.argsort(solution_costs)
print_to = filename[:-4] + "_all_solutions_v4.txt"
count = 1
with open(print_to, mode="a") as file:
file.write("filename: %s \nmissing_weight: %f \nn: %d\n" % (filename, missing_weight, n))
for i in cost_sorted_i:
file.write("%d. best solution with cost %f\n" % (count, solution_costs[i]))
count += 1
for j in range(0,n):
file.write(f"{parents[i, j]} ")
file.write("\n")
def merged_to_file(solutions, costs, filename, missing_weight, n, x, n_merges, output_path):
"""
A function to write the merged solution(s) to a file, named like the input instance ending with _merged.txt.
"""
print_to = output_path + "merged_v4.txt"
with open(print_to, mode="a") as file:
file.write("filename: %s \nmissing_weight: %f \nn: %d \nx (solutions merged): %d\nmerged solutions:\n" % (filename, missing_weight, n, x))
for i in range(n):
file.write(f"{solutions[0, i]} ")
def merged_short_print(solutions, costs, filename, missing_weight, n, x, n_merges):
for j in range(n_merges):
cluster_sizes = {}
for i in range(n):
curr = solutions[j, i]
if curr in cluster_sizes:
cluster_sizes[curr] += 1
else:
cluster_sizes[curr] = 1
print(cluster_sizes)
#### merge: scan-variant ####
@njit
def weighted_decision_scan(x, y, connectivity, f_vertex_costs, f_sizes, f_parents):
"""
This function is a helper function for merging functions. It generates a weight for cluster center x and another node y by counting the costs over all solutions for two scenarios:
1: y is in the same cluster as x
0: y is in another cluster
The return value is between -1 and 1, -1 for certainly not connected, 1 for certainly connected. A value of 0 would indicate that connected or not connected would (in mean) yield the same costs (as in: the error is not big enough to make a difference).
"""
sol_len = len(f_parents)
sum_for_0 = 0
sum_for_1 = 0
count_0 = 0
count_1 = 0
for i in range(0,sol_len):
x_cost = f_vertex_costs[i, x]
y_cost = f_vertex_costs[i, y]
if connectivity[i]:
sum_for_1 += x_cost + y_cost
count_1 += 1
else:
sum_for_0 += x_cost + y_cost
count_0 += 1
if count_0 > 0:
cost_0 = sum_for_0/count_0
if count_1 > 0:
cost_1 = sum_for_1/count_1
if cost_0 == 0 and cost_1 == 0:
print("Warning: Both together and single get cost 0 - something went wrong!")
else:
return (cost_0 - cost_1) / (cost_0 + cost_1)
else:
# Falls kein Eintrag 1 gehört Knoten recht sicher nicht zum Cluster
return -1.0
else:
# Falls kein Eintrag 0 gehört Knoten recht sicher zum Cluster
return 1.0
# Falls Rückgabe positiv: Entscheidung für 1 (zusammen), falls negativ: Entscheidung für 0 (getrennt).
# Je näher Rückgabewert an 0, desto unsicherer die Entscheidung.
# Falls kein voriger Fall eintritt (Häufigkeit entscheidet/ Verhältnis liegt vor):
return 0.0
def merged_solution_scan(solution_costs, vertex_costs, parents, sizes, missing_weight, n, filename, output_path, union_threshold):
"""
First merge algorithm. It calculates cluster masks for each cluster center:
True, if the node is in the same component with cluster center,
False otherwise.
For these cluster masks, for each cluster center x and each other node y a weighted decision value is calculated. Is this weight better than the previous one, y gets assigned to new cluster center x. X then gets the weight of the maximum weight over all y, except if that is lower than its previous weight. Tree-like structures can emerge in such cases. Those trees are not handled yet, however they indicate a conflict in the solution, as a node that is both child and parent belongs to two distinct clusters.
"""
sol_len = len(solution_costs)
# Neue Lösung als Array anlegen:
merged_sol = np.arange(n) #dtype = np.int64 not supported by numba
merged_sizes = np.ones(n, dtype=np.int64)
# Arrays anlegen für Vergleichbarkeit der Cluster:
connectivity = np.zeros(sol_len, dtype=np.int8) #np.bool not supported
graph_file = open(filename, mode="r")
l = 0
#wd_f = open(output_path + "wd_v4.txt", mode = "a")
for line in graph_file:
l += 1
# Kommentar-Zeilen überspringen
if line[0] == "#":
continue
splitted = line.split()
nodes = np.array(splitted[:-1], dtype=np.int64)
weight = np.float64(splitted[2])
i = nodes[0]
j = nodes[1]
if weight < 0:
continue
# Fülle Cluster-Masken
for x in range(sol_len):
connectivity[x] = np.int8(parents[x, i] == parents[x, j])
# Berechne Zugehörigkeit zu Cluster (bzw. oder Nicht-Zugehörigkeit)
# Alle vorigen Knoten waren schon als Zentrum besucht und haben diesen Knoten daher schon mit sich verbunden (bzw. eben nicht) - Symmetrie der Kosten!
wd = weighted_decision_scan(i, j, connectivity, vertex_costs, sizes, parents)
#wd_f.write(str(l) + " " + str(wd) +"\n")
# Falls Gewicht groß genug:
if wd > union_threshold:
union(i, j, merged_sol, merged_sizes)
#wd_f.close()
result = np.zeros((2,n))
result[0] = merged_sol
result[1] = merged_sizes
return result
#### merge repair variants: with/without scan ####
@njit
def repair_merged_v4_nd(merged, merged_sizes, solution_costs, vertex_costs, parents, sizes, n, node_dgree, big_border):
sol_len = len(solution_costs)
ccs_mndgr = calculate_mean_nodedgr_nd(merged, merged_sizes, node_dgree)
ccs = ccs_mndgr[0]
mean_ndgree = ccs_mndgr[1]
connectivity = np.zeros(sol_len, dtype=np.int8)
for s_center_i in range(len(ccs)):
# s_center soll klein genug sein
s_center = ccs[s_center_i]
if merged_sizes[s_center] > mean_ndgree[s_center_i] * big_border:
continue
# Detektiere und verbinde "Mini-Cluster" (Wurzel des Clusters soll verbunden werden);
# Reparatur wird versucht, wenn die Größe des Clusters weniger als halb so groß ist wie der Knotengrad angibt, dh. die lokale Fehlerrate wäre bei über 50% in der Probleminstanz.
best_fit = s_center
min_mwc = 1.7976931348623157e+308
for b_center_i in range(len(ccs)):
# b_center soll groß genug sein
b_center = ccs[b_center_i]
if merged_sizes[b_center] <= mean_ndgree[b_center_i] * big_border:
continue
# Falls Cluster zusammen deutlich zu groß wären, überspringt diese Kombination direkt
if merged_sizes[s_center] + merged_sizes[b_center] > 1.29 * mean_ndgree[b_center_i]:
continue
for x in range(0,sol_len):
if parents[x, s_center] == parents[x, b_center]:
connectivity[x] = 1
else:
connectivity[x] = 0
# Berechne Gewicht:
mwc = mean_weight_connected(s_center, connectivity, vertex_costs, sizes, parents)
if mwc == -1:
continue
if mwc < min_mwc:
# Aktualisieren von Minimalen Kosten
min_mwc = mwc
best_fit = b_center
# Verbinde das Cluster mit dem Cluster, das im Mittel für s_center am günstigsten ist.
union(s_center, best_fit, merged, merged_sizes)
result = np.zeros((2,n), dtype=np.int64)
result[0] = merged
result[1] = merged_sizes
return result
@njit
def mean_weight_connected(s_center, connectivity, vertex_costs, sizes, parents):
sol_len = len(connectivity)
mwc = 0.0
count = 0
for i in range(sol_len):
if connectivity[i]:
mwc += vertex_costs[i, s_center]
count += 1
if count == 0:
return -1.0
return mwc/count
@njit
def calculate_mean_nodedgr_array(merged, merged_sizes, node_dgree, cluster_centers):
cluster_mean_nodedgr = np.zeros(len(cluster_centers), dtype=np.int64)
for c in range(len(cluster_centers)):
for i in range(len(merged)):
if merged[i] == cluster_centers[c]:
cluster_mean_nodedgr[c] += node_dgree[i]
cluster_mean_nodedgr[c] /= merged_sizes[cluster_centers[c]]
cmn_array = np.zeros(len(merged), dtype=np.int64)
for i in range(len(cluster_centers)):
c = cluster_centers[i]
cmn_array[c] = cluster_mean_nodedgr[i]
return cmn_array
@njit
def calculate_mean_nodedgr_nd(merged, merged_sizes, node_dgree):
cluster_centers = pd.unique(merged)
cluster_mean_nodedgr = np.zeros(len(cluster_centers), dtype=np.int64)
for c in range(len(cluster_centers)):
for i in range(len(merged)):
if merged[i] == cluster_centers[c]:
cluster_mean_nodedgr[c] += node_dgree[i]
cluster_mean_nodedgr[c] /= merged_sizes[cluster_centers[c]]
result = np.zeros((2,len(cluster_centers)), dtype=np.int64)
result[0] = cluster_centers
result[1] = cluster_mean_nodedgr
return result
def repair_merged_v4_scan(merged, merged_sizes, solution_costs, vertex_costs, parents, sizes, n, node_dgree, big_border, filename):
sol_len = len(solution_costs)
cluster_centers = pd.unique(merged)
mean_ndgree = calculate_mean_nodedgr_array(merged, merged_sizes, node_dgree, cluster_centers)
connectivity = np.zeros(sol_len, dtype=np.int8)
best_fits = np.zeros(n, dtype=np.int64)
min_mwcs = np.zeros(n, dtype = np.float64)
for i in range(n):
best_fits[i] = -1
min_mwcs[i] = 1.7976931348623157e+308
graph_file = open(filename, mode="r")
for line in graph_file:
# Kommentar-Zeilen überspringen
if line[0] == "#":
continue
splitted = line.split()
nodes = np.array(splitted[:-1], dtype=np.int64)
weight = np.float64(splitted[2])
i = nodes[0]
j = nodes[1]
# Nur positive Kanten berücksichtigen
if weight < 0:
continue
#Clusterzentren ermitteln
s_center = merged[i]
b_center = merged[j]
# ggf. Benennung ändern (b: big, s: small)
if merged_sizes[s_center] > merged_sizes[b_center]:
tmp = s_center
s_center = b_center
b_center = tmp
# Clustergrößen ermitteln
s_center_s = merged_sizes[s_center]
b_center_s = merged_sizes[b_center]
if b_center_s < big_border * mean_ndgree[b_center]:
continue
if s_center_s >= big_border * mean_ndgree[s_center]:
continue
if s_center_s + b_center_s > 1.29 * mean_ndgree[s_center]:
continue
if s_center_s + b_center_s > 1.29 * mean_ndgree[b_center]:
continue
for x in range(0,sol_len):
if parents[x, i] == parents[x, j]:
connectivity[x] = 1
else:
connectivity[x] = 0
# Berechne Gewicht:
mwc = mean_weight_connected(s_center, connectivity, vertex_costs, sizes, parents)
if mwc == -1:
continue
if mwc < min_mwcs[s_center]:
# Aktualisieren von Minimalen Kosten
min_mwcs[s_center] = mwc
best_fits[s_center] = b_center
# Laufe über alle großen Cluster (denen kleine zugewiesen wurden) und verbinde diese mit den günstigsten Kandidaten,
# bis das Cluster (deutlich) zu voll wäre.
bf_unique = | pd.unique(best_fits) | pandas.unique |
"""
Open Power System Data
Household Datapackage
validation.py : fix possible errors and wrongly measured data.
"""
import logging
logger = logging.getLogger(__name__)
import os
import yaml
import pytz
import numpy as np
import pandas as pd
from datetime import datetime, timedelta
from .tools import update_progress, derive_power
def validate(household, household_data, config_dir='conf', verbose=False):
'''
Search for measurement faults in several data series of a DataFrame and remove them
Parameters
----------
household : dict
Configuration dictionary of the household
household_data : pandas.DataFrame
DataFrame to inspect and possibly fix measurement errors
config_dir : str
directory path where all configurations can be found
output : boolean
Flag, if the validated feeds should be printed as human readable CSV files
Returns
----------
result: pandas.DataFrame
Adjusted DataFrame with result series
'''
result = pd.DataFrame()
logger.info('Validate %s series', household['name'])
feeds_columns = household_data.columns.get_level_values('feed')
feeds_configs = _read_adjustments(household['id'], config_dir)
feeds_output = pd.DataFrame()
feeds_existing = len(household_data.columns)
feeds_success = 0
for feed_name in household['series'].keys():
feed = household_data.loc[:, feeds_columns==feed_name].dropna()
#Take specific actions, depending on one-time occurrences for the specific feed
if feed_name in feeds_configs:
for feed_configs in feeds_configs[feed_name]:
feed = _series_adjustment(feed_configs, feed, feed_name)
# Keep only the rows where the energy values are within +3 to -3 times the standard deviation.
error_std = np.abs(feed - feed.mean()) > 3*feed.std()
if np.count_nonzero(error_std) > 0:
logger.debug("Deleted %s %s values: %s energy values 3 times the standard deviation",
household['name'], feed_name, str(np.count_nonzero(error_std)))
# Keep only the rows where the energy values is increasing
feed_fixed = feed[~error_std]
error_inc = feed_fixed < feed_fixed.shift(1)
feed_size = len(feed_fixed.index)
for time in error_inc.replace(False, np.NaN).dropna().index:
i = feed_fixed.index.get_loc(time)
if i > 2 and i < feed_size:
error_flag = None
# If a rounding or transmission error results in a single value being too big,
# fix that single data point, else flag all decreasing values
if feed_fixed.iloc[i,0] >= feed_fixed.iloc[i-2,0] and not error_inc.iloc[i-2,0]:
error_inc.iloc[i,0] = False
error_inc.iloc[i-1,0] = True
elif all(feed_fixed.iloc[i-1,0] > feed_fixed.iloc[i:min(feed_size-1,i+10),0]):
error_flag = feed_fixed.index[i]
else:
j = i+1
while j < feed_size and feed_fixed.iloc[i-1,0] > feed_fixed.iloc[j,0]:
if error_flag is None and j-i > 10:
error_flag = feed_fixed.index[i]
error_inc.iloc[j,0] = True
j = j+1
if error_flag is not None:
logger.warn('Unusual behaviour at index %s for %s: %s', error_flag.strftime('%d.%m.%Y %H:%M'), household['name'], feed_name)
if np.count_nonzero(error_inc) > 0:
feed_fixed = feed_fixed[~error_inc]
logger.debug("Deleted %s %s values: %s decreasing energy values",
household['name'], feed_name, str(np.count_nonzero(error_inc)))
# Notify about rows where the derived power is significantly larger than the standard deviation value
feed_power = derive_power(feed_fixed)
quantile = feed_power[feed_power > 0].quantile(.99)[0]
error_qnt = (feed_power.abs() > 3*quantile).shift(-1).fillna(False)
error_qnt.columns = feed_fixed.columns
if np.count_nonzero(error_qnt) > 0:
feed_fixed = feed_fixed[~error_qnt]
logger.debug("Deleted %s %s values: %s power values 3 times .99 standard deviation",
household['name'], feed_name, str(np.count_nonzero(error_qnt)))
if not feed_fixed.empty:
# Always begin with an energy value of 0
feed_fixed -= feed_fixed.dropna().iloc[0,0]
if verbose:
os.makedirs("raw_data", exist_ok=True)
error_std = error_std.replace(False, np.NaN)
error_inc = error_inc.replace(False, np.NaN)
error_qnt = error_qnt.replace(False, np.NaN)
feed_columns = [feed_name+"_energy", feed_name+"_power", feed_name+'_error_std', feed_name+'_error_inc', feed_name+'_error_qnt']
feed_csv = pd.concat([feed, derive_power(feed), error_std, error_inc, error_qnt], axis=1)
feed_csv.columns = feed_columns
feed_csv.to_csv(os.path.join("raw_data", household['id']+'_'+feed_name+'.csv'),
sep=',', decimal='.', encoding='utf-8')
feed_output = pd.concat([feed_fixed, derive_power(feed_fixed), error_std, error_inc, error_qnt], axis=1)
feed_output.columns = feed_columns
feeds_output = | pd.concat([feeds_output, feed_output], axis=1) | pandas.concat |
from elasticsearch import Elasticsearch, helpers
import pandas
from utils.log import *
class ES(object):
def __init__(self, conf):
self.es = Elasticsearch([conf['es_host']])
self.winlogbeat = conf['winlogbeat_index']
def insert_behaviors(self, _index, data):
records = []
for _be in data:
_cols = _be.get_attribute_names()
_record = _be.serialize()
try:
records.append({
'_index': _index + _be.getname().lower(),
'_type': 'behavior',
'_source': pandas.DataFrame([_record], columns=_cols).loc[0].to_json(default_handler=str),
})
except Exception as e:
log_error('Elasticsearch insert failed. {}'.format(e))
log_error(pandas.DataFrame([_record], columns=_cols).loc[0])
helpers.bulk(self.es, records)
def query(self, _index, doctype, qstring):
records = []
try:
page = self.es.search(index=_index, doc_type=doctype, scroll='2m', size=1000, q=qstring, timeout='10m')
sid = page['_scroll_id']
scroll_size = page['hits']['total']
docs = page['hits']['hits']
records += [x['_source'] for x in docs]
while scroll_size > 0:
page = self.es.scroll(scroll_id=sid, scroll='5m')
sid = page['_scroll_id']
scroll_size = len(page['hits']['hits'])
docs = page['hits']['hits']
records += [x['_source'] for x in docs]
return | pandas.DataFrame(records) | pandas.DataFrame |
import pandas as pd
# from WindPy import *
import sympy as smp
import scipy as scp
import scipy.stats as sss
import scipy.optimize as sop
import numpy as np
import pandas as pd
from datetime import datetime, date, timedelta
import matplotlib as mpl
import matplotlib.pyplot as plt
from fh_tools.fh_utils import get_cache_file_path
from scipy.stats import norm
import matplotlib.mlab as mlab
import pymysql
import json
from config_fh import get_db_engine, ANALYSIS_CACHE_FILE_NAME, get_db_session, get_redis, STR_FORMAT_DATE
zhfont1 = mpl.font_manager.FontProperties(fname='C:\Windows\Fonts\simkai.ttf')
def copula_func(copula_family, Xs_in, alpha_in):
Num_vars_in = len(Xs_in)
inner_fun = 0
if copula_family == 'Gumbel':
for i in range(Num_vars_in):
inner_fun += (-smp.log(Xs_in[i])) ** alpha_in
gumbel_expr = smp.exp(-inner_fun ** (1 / alpha_in))
return gumbel_expr
elif copula_family == 'Clayton':
for i in range(Num_vars_in):
inner_fun += Xs_in[i] ** (-alpha_in)
clayton_expr = (inner_fun - Num_vars_in + 1) ** (-1 / alpha_in)
return clayton_expr
else:
print('Copula function not ready yet')
def copula_diff(func_in, Xs_in):
Num_vars_in = len(Xs_in)
temp_func = func_in.copy()
for i in range(Num_vars_in):
temp_func = smp.diff(temp_func, Xs_in[i])
return temp_func
def estimate_parameter(diff_func, data_in, Xs_in, alpha_in):
temp = 1
for i in range(len(data_in)):
temp = temp * diff_func.subs([(Xs_in[j], data_in[i, j]) for j in range(len(Xs_in))])
myfunc = smp.lambdify(alpha_in, -smp.log(temp), 'math')
##function used to make alpha as para, x as value only
alpha_num = sop.fminbound(myfunc, 1, 10)
return alpha_num
def rnd_generator(alpha_in, var_num_in, size_in):
temp = np.zeros((size_in, var_num_in))
for i in range(size_in):
gamma_rv = np.random.gamma(1, 1 / alpha_in)
rnd_n = np.random.rand(var_num_in)
rnd = (1 - 1 / gamma_rv * np.log(rnd_n)) ** (-1 / alpha_in)
temp[i, :] = rnd
return temp
def cal_maxdd(nvlist):
dd = []
for nvindex, nv in enumerate(nvlist):
nvmax = max(nvlist[0:nvindex + 1])
dd_now = nv / nvmax - 1
dd.append(dd_now)
return min(dd)
class stress_test:
def __init__(self, copula_family):
self.coupla_family = copula_family
self.sql_getfund = '''select nav_date2, max(nav_acc_max) as nav_acc_mm from (
select adddate(nav_date, 4 - weekday(nav_date)) as nav_date2, max(nav_acc) as nav_acc_max
from fund_nav
where wind_code='%s' and nav_date >= '%s' and nav_date <= '%s' group by nav_date) as nav_t
group by nav_date2 order by nav_date2'''
def get_fund_nv(self, fund_list, start_date, end_date):
fund_nv = pd.DataFrame()
for fund_code in fund_list:
sql_fund = self.sql_getfund % (fund_code, start_date, end_date)
engine = get_db_engine()
fund_nv_tmp = pd.read_sql(sql_fund, engine)
if len(fund_nv) == 0:
fund_nv = fund_nv_tmp
else:
fund_nv = fund_nv.merge(fund_nv_tmp, on='nav_date2')
fund_nv.dropna(how='any', inplace=True)
fund_nv.set_index('nav_date2', inplace=True)
return fund_nv
def simulate_fund_profit(self, fund_nv):
fund_profit = fund_nv.apply(lambda x: x / x.shift(1) - 1)
fund_profit.dropna(how='any', inplace=True)
# print(fund_profit)
cdfs = np.zeros_like(fund_profit)
norm_mean = np.zeros(fund_profit.columns.size)
norm_var = np.zeros(fund_profit.columns.size)
for i in range(fund_profit.columns.size):
norm_mean[i], norm_var[i] = sss.norm.fit(fund_profit.iloc[:, i])
cdfs[:, i] = sss.norm.cdf(fund_profit.iloc[:, i], norm_mean[i], norm_var[i])
# %%
Num_vars = fund_profit.columns.size
Xs = smp.symbols('X1:%d' % (Num_vars + 1))
# print(Num_vars, Xs)
alpha = smp.symbols('alpha')
myfunc = copula_func(self.coupla_family, Xs, alpha)
myfunc_diff = copula_diff(myfunc, Xs)
# print(Num_vars, Xs,1)
alpha_num = estimate_parameter(myfunc_diff, cdfs, Xs, alpha)
# %%
simu_data = rnd_generator(alpha_num, len(Xs), 500)
simu_data_conditional = simu_data[simu_data[:, 0] < 0.1]
simu_real = simu_data.copy()
for i in range(fund_profit.columns.size):
simu_real[:, i] = norm.ppf(simu_real[:, i], norm_mean[i], norm_var[i])
# for i in range(testdata.columns.size):
# simu_data_conditional[:,i]=sss.norm.ppf(simu_data_conditional[:,i], norm_mean[i], norm_var[i])
# print(simu_data)
return simu_real
def get_max_drawdown(self, wind_code_list, start_date, end_date, weight_list, simulate_count):
fnv = self.get_fund_nv(wind_code_list, start_date, end_date)
weight_list = np.array(weight_list)
weight_list = weight_list / sum(weight_list)
max_dd_list = []
for i in range(simulate_count):
simu = self.simulate_fund_profit(fnv)
simu_pd = | pd.DataFrame(simu, columns=wind_code_list) | pandas.DataFrame |
__all__ = [
"read_clock_paramaters",
"read_weather_inputs",
"read_model_parameters",
"read_irrigation_management",
"read_field_management",
"read_groundwater_table",
"compute_variables",
"compute_crop_calander",
"calculate_HIGC",
"calculate_HI_linear",
"read_model_initial_conditions",
"create_soil_profile",
]
# Cell
import numpy as np
import os
import pandas as pd
from .classes import *
import pathlib
from copy import deepcopy
import aquacrop
# Cell
def read_clock_paramaters(SimStartTime, SimEndTime, OffSeason=False):
"""
function to read in start and end simulaiton time and return a `ClockStructClass` object
*Arguments:*\n
`SimStartTime` : `str`: simulation start date
`SimEndTime` : `str` : simulation start date
`OffSeason` : `bool` : simulate off season true, false
*Returns:*
`ClockStruct` : `ClockStructClass` : time paramaters
"""
# extract data and put into numpy datetime format
SimStartTime = pd.to_datetime(SimStartTime)
SimEndTime = pd.to_datetime(SimEndTime)
# create object
ClockStruct = ClockStructClass()
# add variables
ClockStruct.SimulationStartDate = SimStartTime
ClockStruct.SimulationEndDate = SimEndTime
ClockStruct.nSteps = (SimEndTime - SimStartTime).days + 1
ClockStruct.TimeSpan = | pd.date_range(freq="D", start=SimStartTime, end=SimEndTime) | pandas.date_range |
# -*- coding: utf-8 -*-
"""Data structure for validating and efficiently slicing
fixed-length segments of typically multichannel time-series data.
"""
import numpy as np
import pandas as pd
from .errors import FitGridError
from . import tools
class Epochs:
"""Container class used for storing epochs tables and exposing statsmodels.
Parameters
----------
epochs_table : pandas DataFrame
long form dataframe containing epochs with equal indices
time : str
time column name
epoch_id : str
epoch identifier column name
channels : list of str
list of channel names to serve as dependent variables
Returns
-------
epochs : Epochs
epochs object
"""
def __init__(self, epochs_table, time, epoch_id, channels):
# channels must be a list of strings
if not isinstance(channels, list) or not all(
isinstance(item, str) for item in channels
):
raise FitGridError('channels should be a list of strings.')
# all channels must be present as epochs table columns
missing_channels = set(channels) - set(epochs_table.columns)
if missing_channels:
raise FitGridError(
'channels should all be present in the epochs table, '
f'the following are missing: {missing_channels}'
)
if not isinstance(epochs_table, pd.DataFrame):
raise FitGridError('epochs_table must be a Pandas DataFrame.')
# these index columns are required for consistency checks
for item in (epoch_id, time):
if item not in epochs_table.index.names:
raise FitGridError(
f'{item} must be a column in the epochs table index.'
)
# check no duplicate column names in index and regular columns
names = list(epochs_table.index.names) + list(epochs_table.columns)
deduped_names = tools.deduplicate_list(names)
if deduped_names != names:
raise FitGridError('Duplicate column names not allowed.')
# make our own copy so we are immune to modification to original table
table = (
epochs_table.copy().reset_index().set_index(epoch_id).sort_index()
)
assert table.index.names == [epoch_id]
snapshots = table.groupby(time)
# check that snapshots across epochs have equal index by transitivity
prev_group = None
for idx, cur_group in snapshots:
if prev_group is not None:
if not prev_group.index.equals(cur_group.index):
raise FitGridError(
f'Snapshot {idx} differs from '
f'previous snapshot in {epoch_id} index:\n'
f'Current snapshot\'s indices:\n'
f'{cur_group.index}\n'
f'Previous snapshot\'s indices:\n'
f'{prev_group.index}'
)
prev_group = cur_group
if not prev_group.index.is_unique:
dupes = tools.get_index_duplicates_table(table, epoch_id)
raise FitGridError(
f'Duplicate values in {epoch_id} index not allowed:\n{dupes}'
)
# checks passed, set instance variables
self.time = time
self.epoch_id = epoch_id
self.channels = channels
self.table = table
self._snapshots = snapshots
self.epoch_index = tools.get_first_group(snapshots).index.copy()
self.time_index = | pd.Index([time for time, _ in snapshots], name=time) | pandas.Index |
#! /usr/bin/env python3
# -*- coding: utf-8 -*-
# =====================================================================
# Copyright (C) 2018-2019 by Owl Data
# author: <NAME>
# =====================================================================
import requests
import json
import time
import pandas as pd
from pandas.tseries.offsets import MonthEnd, QuarterEnd, YearEnd
from ._owlerror import OwlError
from ._owltime import _DataID
# --------------------
# BLOCK API擷取資料
# --------------------
# 核心程式
class OwlData(_DataID):
def __init__(self, auid:str, ausrt:str):
'''
請輸入個人數據貓頭鷹相關資料,以便登入取得財經數據
Parameters
----------
:param auid: str
- 請輸入數據貓頭鷹 AppID
:param ausrt: str
- 請輸入數據貓頭鷹 應用程式密鑰
'''
self._token = {
'token_url':"https://owl.cmoney.com.tw/OwlApi/auth",
'token_params':"appId=" + auid + "&appSecret=" + ausrt,
'token_headers':{'content-type': "application/x-www-form-urlencoded"}, #POST表單,預設的編碼方式 (enctype)
'data_url':"https://owl.cmoney.com.tw/OwlApi/api/v2/json/",
'testmap':"PYCtrl-14881b",
'pythonmap':"PYCtrl-14882b"
}
# 取得 TOKEN 結果
self._token_result = ''
# Data Token
self._data_headers = {}
# 連線進入並輸出連線狀態
self.status_code = self._request_token_authorization()
# OwlTime Class
super().__init__()
def __repr__(self):
return '貓頭鷹資料庫連線狀態: {}'.format(str(self.status_code))
def _request(self,*args,**kargs):
'''當連線出現異常時,重新啟動下載機制'''
i=3
while i >=0:
try:
return requests.request(*args,**kargs)
except:
time.sleep(3)
i-=1
# Token 取得
def _request_token_authorization(self) -> int:
'''請求權限交換'''
self._token_result = self._request("POST",self._token['token_url'],
data = self._token['token_params'],
headers = self._token['token_headers'])
if (self._token_result.status_code == 200):
token = json.loads(self._token_result.text).get("token")
self._data_headers = {'authorization':'Bearer ' + token}
self._pdid_map()
return self._token_result.status_code
elif(self._token_result.status_code in OwlError._http_error.keys()):
print('錯誤代碼: {} '.format(str(self._token_result.status_code)),OwlError._http_error[self._token_result.status_code])
else:
print("連線錯誤,請洽業務人員")
# 呼叫 OwlData 資料下載
def _data_from_owl(self, url:str) -> pd.DataFrame:
'''
輸入API網址,獲取對應的數據資料
Parameters
------------
:param url: str
- 在url的地方輸入API網址,回傳數據資料 - 個股/多股
Returns
--------
:DataFrame: 輸出分別為個股與多股
Examples
---------
個股/多股: 輸出帶有DataFrame的資料格式
個股:
日期 股票名稱 開盤價 最高價 最低價 ...
0 20171229 台泥 36.20 36.80 36.10 ...
1 20171228 台泥 36.10 36.25 36.00 ...
2 20171227 台泥 36.10 36.45 36.00 ...
3 20171226 台泥 36.10 36.50 35.95 ...
4 20171225 台泥 35.30 36.25 35.10 ...
多股:
股票代號 股票名稱 日期 開盤價 ...
0 1101 台泥 20171229 36.20 ...
1 1102 亞泥 20171229 27.80 ...
2 1103 嘉泥 20171229 12.85 ...
3 1104 環泥 20171229 22.85 ...
Notes
-------
先請求網址回覆200後,才會抓取資料(網址輸入錯誤,才會撈不到資料)
個股: 假設基準日: 20190701、股票代號: 1101、期數: 20,則會獲取自20190701往前20筆 1101的資料
多股: 取指定日期當天,各檔的數據資料
'''
data_result = self._request("GET", url, headers = self._data_headers)
# 若逾時連線,則重新再次連線一次
try:
if data_result.status_code == 200:
pass
except:
data_result = self._request("GET", url, headers = self._data_headers)
# status code 下載判別狀態
try:
if data_result.status_code == 200:
data=json.loads(data_result.text)
return pd.DataFrame(data.get('Data'), columns = data.get('Title'))
elif data_result.status_code in OwlError._http_error.keys():
print('錯誤代碼: {:s} '.format(str(self._token_result.status_code)),
OwlError._http_error[self._token_result.status_code])
return 'error'
except:
return 'error'
# 修正資料
def _check(self, result:pd.DataFrame, freq=None, num_col=2, colists=None, pd_id=None) -> pd.DataFrame:
'''
商品檢查點
Parameters
----------
:param result: DataFrame
- 輸入原始表格
:param freq: str
- 表格頻率
:param num_col: int
- 數值化資料欄位起點
:param colists: list, default None
- 填入欲查看的欄位名稱,未寫輸入則取全部欄位
:param pd_id: str
- 商品代碼
Returns
----------
DataFrame
'''
try:
if result.empty:
print('SidError:',OwlError._dicts["SidError"])
return result
if result is not 'error':
# 日期修正
if freq == 'd':
result['日期'] = [pd.to_datetime(i) if i !='' else '' for i in result['日期']]
if '股票代號' not in result.columns:
result.sort_values('日期', inplace = True)
result.reset_index(drop = True, inplace = True)
elif freq == 'm':
result['年月'] = [pd.to_datetime(i,format='%Y%m')+MonthEnd(1) if i !='' else '' for i in result['年月']]
if '股票代號' not in result.columns:
result.sort_values('年月', inplace = True)
result.reset_index(drop = True, inplace = True)
elif freq == 'q':
result['年季'] = result['年季'].apply(lambda x: x[0:4]+x[4:6].replace('0','Q'))
result['年季'] = [pd.to_datetime(i)+Quar | terEnd(1) | pandas.tseries.offsets.QuarterEnd |
import pandas as pd
import numpy as np
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.preprocessing import MinMaxScaler, StandardScaler
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV
from sklearn.neural_network import MLPRegressor
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import GradientBoostingRegressor, RandomForestRegressor, AdaBoostRegressor
from xgboost import XGBRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.svm import SVR
from warnings import filterwarnings
import tensorflow as tf
from tensorflow import keras
filterwarnings('ignore')
# Data
features = pd.read_csv('dengue_features_train.csv', index_col=[0, 1, ])
label = pd.read_csv('dengue_labels_train.csv', index_col=[0, 1, ])
# SJ and IQ
sj_features, iq_features = features.loc['sj'], features.loc['iq']
sj_label, iq_label = label.loc['sj'], label.loc['iq']
sj_features.drop(columns=['week_start_date'], axis=1, inplace=True)
iq_features.drop(columns=['week_start_date'], axis=1, inplace=True)
# NaN fill
sj_features.fillna(method='ffill', inplace=True)
iq_features.fillna(method='ffill', inplace=True)
# feature engineering
sj_features['station_range_temp_c'] = sj_features['station_max_temp_c'] - \
sj_features['station_min_temp_c']
iq_features['station_range_temp_c'] = iq_features['station_max_temp_c'] - \
iq_features['station_min_temp_c']
sj_features.drop(columns=['station_max_temp_c',
'station_min_temp_c', 'station_avg_temp_c'], axis=1, inplace=True)
iq_features.drop(columns=['station_max_temp_c',
'station_min_temp_c', 'station_avg_temp_c'], axis=1, inplace=True)
sj_features['reanalysis_range_air_temp_k'] = sj_features['reanalysis_max_air_temp_k'] - \
sj_features['reanalysis_min_air_temp_k']
iq_features['reanalysis_range_air_temp_k'] = iq_features['reanalysis_max_air_temp_k'] - \
iq_features['reanalysis_min_air_temp_k']
sj_features.drop(columns=['reanalysis_max_air_temp_k',
'reanalysis_min_air_temp_k', 'reanalysis_tdtr_k', 'reanalysis_avg_temp_k', 'reanalysis_dew_point_temp_k'], axis=1, inplace=True)
iq_features.drop(columns=['reanalysis_max_air_temp_k',
'reanalysis_min_air_temp_k', 'reanalysis_tdtr_k', 'reanalysis_avg_temp_k', 'reanalysis_air_temp_k', 'reanalysis_dew_point_temp_k'], axis=1, inplace=True)
ndvi = ['ndvi_sw', 'ndvi_se', 'ndvi_nw', 'ndvi_ne']
precip = ['station_precip_mm', 'precipitation_amt_mm',
'reanalysis_sat_precip_amt_mm']
sj_ndvi_data, sj_precip_data = sj_features[ndvi], sj_features[precip]
sj_ndvi_data['minimum_ndvi'], sj_precip_data['precip'] = sj_ndvi_data.min(
axis=1), sj_precip_data.max(axis=1)
iq_ndvi_data, iq_precip_data = iq_features[ndvi], iq_features[precip]
iq_ndvi_data['minimum_ndvi'], iq_precip_data['precip'] = iq_ndvi_data.min(
axis=1), iq_precip_data.max(axis=1)
sj_features['minimum_ndvi'], sj_features['precip'] = sj_ndvi_data['minimum_ndvi'].copy(
), sj_precip_data['precip'].copy()
iq_features['minimum_ndvi'], iq_features['precip'] = iq_ndvi_data['minimum_ndvi'].copy(
), iq_precip_data['precip'].copy()
sj_features.drop(columns=ndvi, axis=1, inplace=True)
sj_features.drop(columns=precip, axis=1, inplace=True)
iq_features.drop(columns=ndvi, axis=1, inplace=True)
iq_features.drop(columns=precip, axis=1, inplace=True)
print(sj_features.columns)
# Merge
sj_data = sj_features.merge(sj_label, on=['year', 'weekofyear'])
iq_data = iq_features.merge(iq_label, on=['year', 'weekofyear'])
# iq_data['total_cases'].hist()
# plt.show()
'''
# Seaborn
# print(plt.colormaps())
corr_sj = sj_data.corr()
sns.heatmap(corr_sj, cmap='gist_heat_r', )
plt.xticks(size=7)
plt.yticks(size=7)
plt.show()
corr_iq = iq_data.corr()
sns.heatmap(corr_iq, cmap='gist_heat_r', )
plt.xticks(size=7)
plt.yticks(size=7)
plt.show()
'''
sj_index = np.all(stats.zscore(sj_data) < 3.5, axis=1)
sj_data = sj_data.loc[sj_index]
iq_index = np.all(stats.zscore(iq_data) < 3.5, axis=1)
iq_data = iq_data.loc[iq_index]
# iq_data['total_cases'].hist()
# plt.show()
# Train test split
data_list = [sj_data, iq_data]
for idx in range(len(data_list)):
if idx == 0:
X_sj = sj_data.drop(columns='total_cases', axis=1)
y_sj = sj_data['total_cases']
elif idx == 1:
X_iq = iq_data.drop(columns='total_cases', axis=1)
y_iq = iq_data['total_cases']
sj_x_train, sj_x_test, sj_y_train, sj_y_test = train_test_split(
X_sj, y_sj, test_size=0.2, random_state=42)
iq_x_train, iq_x_test, iq_y_train, iq_y_test = train_test_split(
X_iq, y_iq, test_size=0.2, random_state=42)
print(f'\nSJ X train shape:{sj_x_train.shape} SJ y train shape:{sj_y_train.shape} SJ X test shape:{sj_x_test.shape} SJ y test shape:{sj_y_test.shape}')
print(f'\nIQ X train shape:{iq_x_train.shape} IQ y train shape:{iq_y_train.shape} IQ X test shape:{iq_x_test.shape} IQ y test shape:{iq_y_test.shape}')
# Scaled
x_train = [sj_x_train, iq_x_train]
scaler = StandardScaler()
for idx in range(len(x_train)):
if idx == 0:
sj_scaled = scaler.fit_transform(sj_x_train)
elif idx == 1:
iq_scaled = scaler.fit_transform(iq_x_train)
scaled_data = [sj_scaled, iq_scaled]
# class
'''
class Model:
def training(self, X, y, estimator):
estimator.fit(X, y)
print(estimator)
X_predict = estimator.predict(X)
mae = mean_absolute_error(y, X_predict)
print(f'Training set mean Absolute Error :{mae}')
def validation(self, X, y, estimator):
scores = cross_val_score(
estimator, X, y, cv=10, scoring='neg_mean_absolute_error')
negative_score = -scores
print(f'10 fold validation scores :{negative_score}')
print(f'\n mean :{negative_score.mean()}')
def prediction_test(self, test_X, y_test, estimator):
X_test_predict = estimator.predict(test_X)
mae_test = mean_absolute_error(y_test, X_test_predict)
print(mae_test)
# Models
xg = Model()
xgbsj = XGBRegressor(seed=42, verbosity=0, colsample_bytree=0.9, learning_rate=0.01,
max_depth=7, min_child_weight=1, n_estimators=170, reg_alpha=0.7, subsample=0.9, objective='reg:tweedie')
xgbiq = XGBRegressor(seed=42, verbosity=0, colsample_bytree=0.5, learning_rate=0.01,
max_depth=7, min_child_weight=1, n_estimators=170, reg_alpha=0.1, subsample=0.7, objective='reg:tweedie')
mlp_sj = MLPRegressor(hidden_layer_sizes=(3, 2, 1,),
learning_rate='adaptive', solver='adam', random_state=42, nesterovs_momentum=False,)
mlp_iq = MLPRegressor(hidden_layer_sizes=(9, 5, 1,),
learning_rate='adaptive', solver='adam', random_state=42, nesterovs_momentum=False,)
'''
# NN
model = keras.models.Sequential([keras.layers.Dense(64, activation='softplus', input_shape=sj_scaled.shape[1:]),
keras.layers.Dense(32, activation='softplus'), keras.layers.Dense(1)])
model.compile(loss='mean_absolute_error', optimizer='sgd')
history = model.fit(sj_scaled, sj_y_train, validation_split=0.1, epochs=200)
print(history.history)
histo = pd.DataFrame(history.history)
histo.plot()
plt.grid(True)
plt.show()
for idx in range(len(scaled_data)):
if idx == 0:
print('\nSJ')
sj_train = xg.training(sj_scaled, sj_y_train, mlp_sj)
elif idx == 1:
print('\nIQ')
iq_train = xg.training(iq_scaled, iq_y_train, xgbiq)
for idx in range(len(scaled_data)):
if idx == 0:
print('\nSJ')
sj_val = xg.validation(sj_scaled, sj_y_train, mlp_sj)
elif idx == 1:
print('\nIQ')
iq_val = xg.validation(iq_scaled, iq_y_train, xgbiq)
'''
# GridSearchCV
param = {'max_depth': [3, 5, 7], 'learning_rate': [0.01], 'n_estimators': [170], 'subsample': [
0.7, 0.5, 0.9], 'min_child_weight': [1, 2, 3], 'reg_alpha': [0.7, 0.9, 0.5], 'colsample_bytree': [0.9, 0.5, 0.7], }
grid_search = GridSearchCV(xgb, param_grid=param, cv=10,
scoring='neg_mean_absolute_error', return_train_score=True)
grid_search.fit(iq_scaled, iq_y_train)
print(grid_search.best_params_)
print(grid_search.best_score_)
'''
# Scaling
x_test = [sj_x_test, iq_x_test]
for idx in range(len(x_test)):
if idx == 0:
sj_scaled_test = scaler.fit_transform(sj_x_test)
elif idx == 1:
iq_scaled_test = scaler.fit_transform(iq_x_test)
test_scaled_data = [sj_scaled_test, iq_scaled_test]
for idx in range(len(test_scaled_data)):
if idx == 0:
sj_test_predict = xg.prediction_test(sj_scaled_test, sj_y_test, mlp_sj)
elif idx == 1:
iq_test_predict = xg.prediction_test(iq_scaled_test, iq_y_test, xgbiq)
mlp_sj.fit(sj_scaled, sj_y_train)
mlp_iq.fit(iq_scaled, iq_y_train)
# Dengue feature test
test_features = pd.read_csv('dengue_features_test.csv', index_col=[0, 1, ])
sj_test_features, iq_test_features = test_features.loc['sj'], test_features.loc['iq']
sj_test_features.drop(columns=['week_start_date'], axis=1, inplace=True)
iq_test_features.drop(columns=['week_start_date'], axis=1, inplace=True)
# NaN fill
sj_test_features.fillna(method='ffill', inplace=True)
iq_test_features.fillna(method='ffill', inplace=True)
# feature engineering
sj_test_features['station_range_temp_c'] = sj_test_features['station_max_temp_c'] - \
sj_test_features['station_min_temp_c']
iq_test_features['station_range_temp_c'] = iq_test_features['station_max_temp_c'] - \
iq_test_features['station_min_temp_c']
sj_test_features.drop(columns=['station_max_temp_c',
'station_min_temp_c', 'station_avg_temp_c'], axis=1, inplace=True)
iq_test_features.drop(columns=['station_max_temp_c',
'station_min_temp_c', 'station_avg_temp_c'], axis=1, inplace=True)
sj_test_features['reanalysis_range_air_temp_k'] = sj_test_features['reanalysis_max_air_temp_k'] - \
sj_test_features['reanalysis_min_air_temp_k']
iq_test_features['reanalysis_range_air_temp_k'] = iq_test_features['reanalysis_max_air_temp_k'] - \
iq_test_features['reanalysis_min_air_temp_k']
sj_test_features.drop(columns=['reanalysis_max_air_temp_k',
'reanalysis_min_air_temp_k', 'reanalysis_tdtr_k', 'reanalysis_avg_temp_k', 'reanalysis_dew_point_temp_k'], axis=1, inplace=True)
iq_test_features.drop(columns=['reanalysis_max_air_temp_k',
'reanalysis_min_air_temp_k', 'reanalysis_tdtr_k', 'reanalysis_avg_temp_k', 'reanalysis_air_temp_k', 'reanalysis_dew_point_temp_k'], axis=1, inplace=True)
sj_test_ndvi_data, sj_test_precip_data = sj_test_features[ndvi], sj_test_features[precip]
sj_test_ndvi_data['minimum_ndvi'], sj_test_precip_data['precip'] = sj_test_ndvi_data.min(
axis=1), sj_test_precip_data.max(axis=1)
iq_test_ndvi_data, iq_test_precip_data = iq_test_features[ndvi], iq_test_features[precip]
iq_test_ndvi_data['minimum_ndvi'], iq_test_precip_data['precip'] = iq_test_ndvi_data.min(
axis=1), iq_test_precip_data.max(axis=1)
sj_test_features['minimum_ndvi'], sj_test_features['precip'] = sj_test_ndvi_data['minimum_ndvi'].copy(
), sj_test_precip_data['precip'].copy()
iq_test_features['minimum_ndvi'], iq_test_features['precip'] = iq_test_ndvi_data['minimum_ndvi'].copy(
), iq_test_precip_data['precip'].copy()
sj_test_features.drop(columns=ndvi, axis=1, inplace=True)
sj_test_features.drop(columns=precip, axis=1, inplace=True)
iq_test_features.drop(columns=ndvi, axis=1, inplace=True)
iq_test_features.drop(columns=precip, axis=1, inplace=True)
# Scaled
x_test_features = [sj_test_features, iq_test_features]
for idx in range(len(x_test_features)):
if idx == 0:
sj_test_actual = scaler.fit_transform(sj_test_features)
elif idx == 1:
iq_test_actual = scaler.fit_transform(iq_test_features)
sj_prediction = mlp_sj.predict(sj_test_actual)
iq_prediction = mlp_iq.predict(iq_test_actual)
print(f'\n{sj_prediction.shape} {iq_prediction.shape}')
sj_final = | pd.DataFrame(sj_prediction) | pandas.DataFrame |
import os
import pathlib
import sys
import febrl_data_transform as transform
import pandas as pd
OUTPUT_DATA_DIR = pathlib.Path(__file__).parent / "holdout"
ORIGINALS_DATA_DIR = pathlib.Path(__file__).parent / "holdout" / "originals"
def main():
# Read in FEBRL data with dupes and separate into A/B/true links.
dataset_A = []
dataset_B = []
true_links = []
for filename in [
"febrl_holdout_dupes_light_mod.csv",
"febrl_holdout_dupes_medium_mod.csv",
"febrl_holdout_dupes_heavy_mod.csv",
]:
_df_A, _df_B, _df_true_links = transform.transform_febrl_dataset_with_dupes(
ORIGINALS_DATA_DIR / filename
)
dataset_A.append(_df_A)
dataset_B.append(_df_B)
true_links.append(_df_true_links)
df_A = pd.concat(dataset_A)
df_B = pd.concat(dataset_B)
df_true_links = pd.concat(true_links)
# Read in extra, non-dupe records and split between datasets A and B.
df_extra = transform.transform_febrl_dataset_without_dupes(
ORIGINALS_DATA_DIR / "febrl_holdout_extras.csv"
)
chunk_size = int(df_extra.shape[0] / 2)
df_A = pd.concat([df_A, df_extra.iloc[0:chunk_size]])
df_B_extra = df_extra.iloc[chunk_size:].rename(
columns={"person_id_A": "person_id_B"}
)
df_B = | pd.concat([df_B, df_B_extra]) | pandas.concat |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
## Import packages
import pandas as pd ## necessary data analysis package
import pyam
import nomenclature as nc
import fileinput
import yaml
import os
import sys
if len(sys.argv) <3:
print('python imputdir outputdir')
else:
inputdir=sys.argv[1]
outputfile=sys.argv[2]
# open config file
with open("settingsUploadOutputsPlan4eu.yml","r") as myyaml:
cfg=yaml.load(myyaml,Loader=yaml.FullLoader)
# compute date range
cfg['T'] = pd.date_range(start=pd.to_datetime(cfg['date_start']),end=pd.to_datetime(cfg['date_end']), freq='1H')
#delete output file if already exists
if os.path.exists(outputfile):
os.remove(outputfile)
i=0
# loop on outputfiles:
# TO BE DONE: include scenarised outputs
if cfg['scenarios']>1:
# compute list of files
for file in cfg['listfiles']:
namefile=file[0:-7]
for i in range(cfg['scenarios']):
listfiles.append(namefile+'_Scen'+i+'_OUT.csv')
else:
listfiles=cfg['listfiles']
for file in listfiles:
if file.find('Scen'):
# find scenario number
scen_index=file[0:-8].split('Scen')[1]
scenario=cfg['scenario']+'|'+scen_index
listscenarios.append(scenario)
else:
scenario=cfg['scenario']
print('writing '+file)
variable=cfg['listfiles'][file]
print(variable)
data= | pd.read_csv(inputdir+'\\'+file) | pandas.read_csv |
#!/usr/bin/env python
import os
import pdb
import glob
import sys
import shutil
import json
import re
import nibabel as nib
from argparse import ArgumentParser
import pandas as pd
import numpy as np
import nilearn.plotting as plotting
import itertools
import matplotlib.colors as colors
import seaborn as sns
import matplotlib.pyplot as plt
import math
from statannot import add_stat_annotation
import bct
import statsmodels
import statsmodels.api as sm
from statsmodels.formula.api import ols
sys.path.insert(0,'..')
import ESM_utils as esm
from scipy import stats
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
## look at mutation differences
def plot_aggregate_roi_performance(ref_pattern_df, pred_pattern_df, roi_labels, output_dir):
plt.figure(figsize=(5,5))
ref_roi_avg = np.mean(ref_pattern_df.loc[:, roi_labels], axis=0)
pred_roi_avg = np.mean(pred_pattern_df.loc[:, roi_labels], axis=0)
sns.regplot(ref_roi_avg, pred_roi_avg, color="indianred")
r,p = stats.pearsonr(ref_roi_avg, pred_roi_avg)
r2 = r ** 2
xmin = np.min(ref_roi_avg)
ymax = np.max(pred_roi_avg)
plt.text(xmin, ymax, "$r^2$ = {0}".format(np.round(r2, 2), fontsize=16))
plt.xlim([-0.01,ymax+.1])
plt.ylim([-0.01,ymax+.1])
plt.xticks(x=16)
plt.yticks(y=16)
plt.xlabel(r"Observed A$\beta$ Probabilities", fontsize=16)
plt.ylabel(r"Predicted A$\beta$ Probabilities", fontsize=16)
plt.title(r"Average A$\beta$ Pattern Across All MC", fontsize=16)
output_path = os.path.join(output_dir, "aggreggate_roi_performance_xsec.png")
plt.tight_layout()
plt.savefig(output_path)
def plot_aggregate_roi_performance_across_eyo(ref_pattern_df, pred_pattern_df, roi_labels, output_dir):
#threshold = 0.3
age_ranges = [[-20, -10], [-10,0], [0,10], [10,20]]
n_cols = len(age_ranges)
fig = plt.figure(figsize=(8,8))
for idx, ar in enumerate(age_ranges):
#loc = 2*(idx+1)-1
ref = np.mean(ref_pattern_df[(ref_pattern_df.DIAN_EYO > ar[0]) & (ref_pattern_df.DIAN_EYO < ar[1])][roi_labels], axis=0)
pred = np.mean(pred_pattern_df[(pred_pattern_df.DIAN_EYO > ar[0]) & (pred_pattern_df.DIAN_EYO < ar[1])][roi_labels], axis=0)
n = ref_pattern_df[(ref_pattern_df.DIAN_EYO > ar[0]) & (ref_pattern_df.DIAN_EYO < ar[1])].shape[0]
r,p = stats.pearsonr(ref, pred)
r2 = r**2
ax1 = plt.subplot(2, 2, idx+1)
sns.regplot(ref, pred, color="indianred")
plt.xlim([-0.1, 1.1])
plt.ylim([-0.1, 1.1])
plt.text(s=r"$r^2$ = {0}".format(np.round(r2, 2)), x=0, y=.9, fontsize=18)
if idx < 2:
plt.xticks([])
else:
plt.xticks(fontsize=18)
plt.yticks(fontsize=18)
plt.title("EYO: {0} to {1} (n = {2})".format(str(ar[0]), str(ar[1]), str(n)), fontsize=18)
fig.text(0.2, 0, r"Observed A$\beta$ Probabilities", fontsize=26)
fig.text(0.01, 0.2, r"Predicted A$\beta$ Probabilities", fontsize=26, rotation=90)
#fig.text(0.2, .95, "Group-level ESM Performance", fontsize=26)
output_path = os.path.join(output_dir, "aggreggate_roi_performance_xsec_across_eyo.png")
#plt.tight_layout()
plt.savefig(output_path)
def roi_performance_hist(ref_pattern_df, pred_pattern_df, roi_labels, output_dir):
roi_r2 = []
for roi in roi_labels:
r = stats.pearsonr(ref_pattern_df.loc[:, roi], pred_pattern_df.loc[:, roi])[0]
roi_r2.append(r**2)
roi_r2_df = pd.DataFrame(columns=["ROI", "r2"])
roi_r2_df['ROI'] = roi_labels
roi_r2_df['r2'] = roi_r2
g = sns.catplot(x='ROI', y='r2',data=roi_r2_df, ci=None,
order = roi_r2_df.sort_values('r2',ascending=False)['ROI'],
kind='bar')
g.set_xticklabels(rotation=90)
g.fig.set_size_inches((14,6))
plt.title('R2 Per ROI Across All Subjects')
output_path = os.path.join(output_dir, "roi_performance_hist.png")
plt.tight_layout()
plt.savefig(output_path)
def plot_conn_matrices(sc_mat, fc_mat):
fig = plt.figure(figsize=(12,8))
ax1 = plt.subplot(1,2,1)
plotting.plot_matrix(sc_mat[0:78,0:78], colorbar=False, cmap="Reds", axes=ax1)
ax1.set_title("Structural Connectivity", fontsize=20)
ax2 = plt.subplot(1,2,2)
plotting.plot_matrix(fc_mat[0:78,0:78], colorbar=True, cmap="Reds", axes=ax2)
ax2.set_title("Functional Connectivity", fontsize=20)
plt.savefig("../../figures/conn_matrices_plot.png")
def plot_subject_performance(res, epicenter, dataset, output_dir):
plt.figure(figsize=(5,5))
pal = {"No": "mediumblue", "Yes": "red"}
face_pal = {"No": "cornflowerblue", "Yes": "indianred"}
g = sns.boxplot(x="mutation_type", y="model_r2", data=res, hue="AB_Positive", palette=face_pal, fliersize=0)
sns.stripplot(x="mutation_type", y="model_r2", data=res, hue="AB_Positive", jitter=True,
split=True, linewidth=0.5, palette=pal)
g.set(xticklabels=["PSEN1", "PSEN2", "APP"])
g.set_ylim([0,.8])
plt.xlabel("Mutation Type")
plt.ylabel("Within subject r2")
if epicenter == "cortical":
title = "Epicenter: MOF, PC, Precuneus"
elif epicenter == "subcortical":
title = "Epicenter: Caudate and Putamen"
elif epicenter == "pc-cac":
title = "Epicenter: Posterior cingulate & Caudal Anterior Cingulate"
plt.title(title)
output_path = os.path.join(output_dir, "within_subject_xsec_perform_across_muttypes.png")
# Get the handles and labels. For this example it'll be 2 tuples
# of length 4 each.
handles, labels = g.get_legend_handles_labels()
# When creating the legend, only use the first two elements
# to effectively remove the last two.
l = plt.legend(handles[0:2], labels[0:2], bbox_to_anchor=(.75, .98), loc=2, borderaxespad=0., title=r"A$\beta$ Positive")
#plt.tight_layout()
plt.savefig(output_path)
def set_ab_positive(ref_pattern_df, rois_to_analyze):
for sub in ref_pattern_df.index:
avg_ab_val = np.mean(list(ref_pattern_df.loc[sub, rois_to_analyze]))
ref_pattern_df.loc[sub, 'PUP_ROI_AB_Mean'] = avg_ab_val
if avg_ab_val > 0.1:
ref_pattern_df.loc[sub, 'AB_Positive'] = "Yes"
else:
ref_pattern_df.loc[sub, 'AB_Positive'] = "No"
return ref_pattern_df
def get_mutation_type(subs, genetic_df):
mutation_type = []
for sub in subs:
mt = genetic_df[(genetic_df.IMAGID == sub)].MUTATIONTYPE.values[0]
mutation_type.append(mt)
return mutation_type
def get_eyo(subs, visit_labels, clinical_df):
eyos = []
for i, sub in enumerate(subs):
eyo = clinical_df[(clinical_df.IMAGID == sub) & (clinical_df.visit == visit_labels[i])].DIAN_EYO.values[0]
eyos.append(eyo)
return eyos
def get_pup_cortical_analysis_cols(roi_labels):
pup_cortical_rois = []
for i, roi in enumerate(roi_labels):
if "precuneus" in roi.lower() or "superior frontal" in roi.lower() \
or "rostral middle frontal" in roi.lower() or "lateral orbitofrontal" in roi.lower() \
or "medial orbitofrontal" in roi.lower() or "superior temporal" in roi.lower() \
or "middle temporal" in roi.lower():
pup_cortical_rois.append(roi)
return pup_cortical_rois
def get_clinical_status(res, clinical_df):
for sub in res.index:
visit = res.loc[sub, 'visit_label']
cdr = clinical_df[(clinical_df.IMAGID == sub) & (clinical_df.visit == visit)].cdrglob.values[0]
if cdr > 0:
res.loc[sub, 'Symptomatic'] = "Yes"
else:
res.loc[sub, 'Symptomatic'] = "No"
res.loc[sub, 'CDR'] = cdr
return res
def plot_effective_anat_dist_vs_ab(ref_pattern_df, acp_matrix, epicenters_idx, roi_labels, output_dir):
# set diagonal to 1
for i in range(0, len(acp_matrix)):
acp_matrix[i][i] = 1
acp_matrix_reciprocal = np.reciprocal(acp_matrix)
acp_effective_dist = bct.distance_wei(acp_matrix_reciprocal)
roi_labels_not_seed = []
effective_anat_distance_dict = {}
for i, roi in enumerate(roi_labels):
dist = 0
for j in epicenters_idx:
dist += acp_effective_dist[0][i, j]
dist = dist / len(epicenters_idx)
#print("{0}: {1}".format(roi, str(np.round(dist,3))))
effective_anat_distance_dict[roi] = dist
roi_dist_ab_df = pd.DataFrame(columns=["Avg_Deposition_Asymptomatic",
"Avg_Deposition_Symptomatic",
"Effective_Anat_Distance"],
index=roi_labels,
dtype="float")
for i,roi in enumerate(roi_labels):
roi_dist_ab_df.loc[roi, "Effective_Anat_Distance"] = effective_anat_distance_dict[roi]
roi_dist_ab_df.loc[roi, "Avg_Deposition_Asymptomatic"] = np.mean(ref_pattern_df[ref_pattern_df.Symptomatic == "No"].loc[:, roi])
roi_dist_ab_df.loc[roi, "Avg_Deposition_Symptomatic"] = np.mean(ref_pattern_df[ref_pattern_df.Symptomatic == "Yes"].loc[:, roi])
fig, (ax1, ax2) = plt.subplots(ncols=2, sharey=False, sharex=False, figsize=(6,3))
axes = [ax1, ax2]
for i, status in enumerate(["Avg_Deposition_Asymptomatic", "Avg_Deposition_Symptomatic"]):
axes[i] = sns.regplot(x="Effective_Anat_Distance", y=status, data=roi_dist_ab_df.loc[roi_labels,:], ax=axes[i])
r, p = stats.pearsonr(roi_dist_ab_df.loc[roi_labels, "Effective_Anat_Distance"],
roi_dist_ab_df.loc[roi_labels, status])
title = status.split("_")[-1]
axes[i].set_xlabel("Effective Anatomical Distance", fontsize=10, axes=axes[i])
axes[i].set_ylabel(r"Regional A$\beta$ Probability", fontsize=10, axes=axes[i])
axes[i].set_title(title, fontsize=10)
axes[i].set_ylim([-0.1,1])
axes[i].text(x=1.5, y=0.8, s="r: {0}\np < 0.01".format(str(np.round(r,3))))
plt.tight_layout()
plt.savefig(os.path.join(output_dir, "effective_anat_dist_vs_ab.png"))
def plot_clinical_status_vs_esm_params(res, output_dir):
plt.figure(figsize=(10,13))
nrows = 2
ncols = 2
params = ["BETAS_est", "DELTAS_est", "BDR_log", "PUP_ROI_AB_Mean"]
face_pal = {"No": "cornflowerblue", "Yes": "indianred"}
titles = ["Production Rate", "Clearance Rate", "Prod/Clear Ratio (Log)", "Amyloid Beta"]
for i, param in enumerate(params):
j = i + 1
plt.subplot(nrows, ncols, j)
g = sns.boxplot(x="Symptomatic", y=param, data=res[res.AB_Positive == "Yes"], palette=face_pal)
add_stat_annotation(g, data=res[res.AB_Positive == "Yes"], x="Symptomatic", y=param,
box_pairs=[("Yes","No")],
test='t-test_ind', text_format='star', loc='inside', verbose=2,
fontsize=18)
plt.ylabel("")
plt.title(titles[i], fontsize=22)
plt.xticks(fontsize=18)
plt.yticks(fontsize=18)
plt.xlabel("Symptomatic",fontsize=18)
plt.tight_layout()
plt.savefig(os.path.join(output_dir, "clinical_status_vs_esm_params.png"))
def model_performance_summary(filename, ref_pattern_df, pred_pattern_df, roi_cols):
ref_region = filename.split("ref-")[1].split("_")[0]
epicenter = filename.split("epicenter-")[1].split("_")[0]
if epicenter.isnumeric():
i = int(epicenter) - 1
epicenter = roi_cols[i]
global_r, global_p = stats.pearsonr(ref_pattern_df[roi_cols].mean(0), pred_pattern_df[roi_cols].mean(0))
global_r2 = np.round(global_r ** 2,2)
sub_r2 = []
for sub in ref_pattern_df.index:
r,p = stats.pearsonr(ref_pattern_df.loc[sub, roi_cols], pred_pattern_df.loc[sub, roi_cols])
r2 = r ** 2
sub_r2.append(r2)
sub_r2_avg = np.round(np.mean(sub_r2),2)
perform_df = pd.DataFrame(columns=["Pipeline", "ReferenceRegion", "Epicenter", "Global_R2", "Subject_R2"])
perform_df.loc[0] = ["PUP", ref_region, epicenter, global_r2, sub_r2_avg]
output_file = os.path.join("../results_csv", filename + ".csv")
perform_df.to_csv(output_file)
def plot_ref_vs_pred_group_brain(ref_pattern_df, pred_pattern_df, roi_labels, output_dir):
dkt_atlas = nib.load("../../data/atlases/dkt_atlas_1mm.nii.gz")
dkt_data = dkt_atlas.get_data()
age_ranges = [[-20, -10], [-10,0], [0,10], [10,20]]
img_slice = [-2, 2, 24]
n_rows = len(age_ranges)
fig = plt.figure(figsize=(20,20))
threshold = 0.1
for idx, ar in enumerate(age_ranges):
loc = 2*(idx+1)-1
curr_ref_probs = np.zeros_like(dkt_data)
curr_pred_probs = np.zeros_like(dkt_data)
for i in range(0, len(roi_labels)):
n = len(ref_pattern_df[(ref_pattern_df.DIAN_EYO > ar[0]) & (ref_pattern_df.DIAN_EYO < ar[1])][roi_labels[i]])
curr_ref_probs[dkt_data==(i+1)] = np.mean(ref_pattern_df[(ref_pattern_df.DIAN_EYO > ar[0]) & (ref_pattern_df.DIAN_EYO < ar[1])][roi_labels[i]])
curr_pred_probs[dkt_data==(i+1)] = np.mean(pred_pattern_df[(pred_pattern_df.DIAN_EYO > ar[0]) & (pred_pattern_df.DIAN_EYO < ar[1])][roi_labels[i]])
curr_ref_probs_img = nib.Nifti1Image(curr_ref_probs, affine=dkt_atlas.affine, header=dkt_atlas.header)
curr_pred_probs_img = nib.Nifti1Image(curr_pred_probs, affine=dkt_atlas.affine, header=dkt_atlas.header)
ax1 = plt.subplot(n_rows, 2, loc)
plotting.plot_stat_map(curr_ref_probs_img, colorbar=True, draw_cross=False, vmax=1, cut_coords=img_slice, axes=ax1, threshold=threshold, cmap="Spectral_r")
plt.title("EYO: {0} to {1} (n = {2})".format(str(ar[0]), str(ar[1]), str(n)), fontsize=36)
ax2 = plt.subplot(n_rows, 2, loc+1)
plotting.plot_stat_map(curr_pred_probs_img, colorbar=True, draw_cross=False, vmax=1, cut_coords=img_slice, axes=ax2, threshold=threshold, cmap="Spectral_r")
fig.text(x=0.25, y=0.9,s="Observed", fontsize=36)
fig.text(x=0.65, y=0.9,s="Predicted", fontsize=36)
plt.savefig(os.path.join(output_dir, "ref_vs_pred_group_brain.png"))
def plot_sub_level_r2_hist(res, output_dir):
plt.figure(figsize=(5,5))
avg = np.mean(res.model_r2)
g = sns.distplot(res.model_r2)
ymin, ymax = g.get_ylim()
xmin, xmax = g.get_xlim()
g.text(x=xmax-0.5, y=ymax-0.5, s="avg: {0}".format(avg))
plt.savefig(os.path.join(output_dir, "sub_level_r2_hist.png"))
def plot_pup_ab_vs_r2(res, output_dir):
fig = plt.figure(figsize=(5,3))
g = sns.lmplot(x="PUP_ROI_AB_Mean", y="model_r2", data=res, lowess=True)
plt.xlabel(r"Average Cortical A$\beta$ Deposition")
plt.ylabel("Within subject r2")
g.set(ylim=(0, 1))
plt.savefig(os.path.join(output_dir, "pup_ab_vs_r2.png"))
def add_csf_biomarker_info(res, biomarker_df, clinical_df):
for sub in res.index:
visit = res.loc[sub, 'visit_label']
csf_ab42 = biomarker_df[(biomarker_df.IMAGID == sub) & (biomarker_df.visit == visit)].CSF_xMAP_AB42.values[0]
csf_tau = biomarker_df[(biomarker_df.IMAGID == sub) & (biomarker_df.visit == visit)].CSF_xMAP_tau.values[0]
csf_ptau = biomarker_df[(biomarker_df.IMAGID == sub) & (biomarker_df.visit == visit)].CSF_xMAP_ptau.values[0]
res.loc[sub, 'CSF_AB42'] = csf_ab42
res.loc[sub, 'CSF_Tau'] = csf_tau
res.loc[sub, 'CSF_pTau'] = csf_ptau
res.loc[sub, 'Age'] = clinical_df[(clinical_df.IMAGID == sub) & (clinical_df.visit == visit)].VISITAGEc.values[0]
res.loc[sub, 'Gender'] = clinical_df[(clinical_df.IMAGID == sub) & (clinical_df.visit == visit)].gender.values[0]
res.loc[sub, 'Education'] = clinical_df[(clinical_df.IMAGID == sub) & (clinical_df.visit == visit)].EDUC.values[0]
return res
def plot_anova_csf_results(res, output_dir):
sns.set_style("white", {'axes.grid' : False})
csf_ab42_formula = 'CSF_AB42 ~ BETAS_est + DELTAS_est + Onset_age + C(Gender) + Age + Education'
csf_ab42_model = ols(csf_ab42_formula, res).fit()
csf_ab42_table = statsmodels.stats.anova.anova_lm(csf_ab42_model, typ=2)
csf_tau_formula = 'CSF_Tau ~ BETAS_est + DELTAS_est + Onset_age + C(Gender) + Age + Education'
csf_tau_model = ols(csf_tau_formula, res).fit()
csf_tau_table = statsmodels.stats.anova.anova_lm(csf_tau_model, typ=2)
csf_ptau_formula = 'CSF_pTau ~ BETAS_est + DELTAS_est + Onset_age + C(Gender) + Age + Education'
csf_ptau_model = ols(csf_ptau_formula, res).fit()
csf_ptau_table = statsmodels.stats.anova.anova_lm(csf_ptau_model, typ=2)
anova_results = {'csf_ab42': {}, 'csf_tau': {}, 'csf_ptau': {}}
csf_types = ['csf_ab42', 'csf_tau', 'csf_ptau']
for i, tbl in enumerate([csf_ab42_table,csf_tau_table,csf_ptau_table]):
csf_type = csf_types[i]
for j, x in enumerate(tbl[tbl.index != 'Residual'].index):
anova_results[csf_type][x] = {}
var_explained = np.round((tbl.loc[x, 'sum_sq'] / np.sum(tbl.loc[:, "sum_sq"])) * 100, 2)
p_value = tbl.loc[x, 'PR(>F)']
var_sig = str(var_explained) + " (" + format(p_value, '.02e') + "), F-value" + str(np.round(tbl.loc[x, 'F'], 3))
anova_results[csf_type][x]['var_explained'] = var_explained
anova_results[csf_type][x]['p_value'] = np.round(p_value,3)
anova_results[csf_type][x]['F_value'] = np.round(tbl.loc[x, 'F'], 3)
anova_barplot_df = | pd.DataFrame(columns=["BETAS_est", "DELTAS_est", "Onset_age", "Age", "Education", "C(Gender)"], index=["CSF_AB42", "CSF_Tau", "CSF_pTau"]) | pandas.DataFrame |
#!/usr/bin/env python
import numpy as np
import pandas as pd
import pytest
from modnet.preprocessing import get_cross_nmi
from modnet.preprocessing import nmi_target
def test_nmi_target():
# Test with linear data (should get 1.0 mutual information, or very close due to algorithm used
# in mutual_info_regression)
npoints = 31
x = np.linspace(0.5, 3.5, npoints)
y = 2*x - 2
z = 4*x + 2
df_feat = pd.DataFrame({'x': x, 'y': y})
df_target = pd.DataFrame({'z': z})
# Here we fix the number of neighbors for the call to sklearn.feature_selection's mutual_info_regression to 2 so
# that we get exactly 1 for the mutual information.
df_nmi_target = nmi_target(df_feat=df_feat, df_target=df_target, n_neighbors=2)
assert df_nmi_target.shape == (2, 1)
assert df_nmi_target.loc['x']['z'] == pytest.approx(1.0)
assert df_nmi_target.loc['y']['z'] == pytest.approx(1.0)
# Same data shuffled
# Shuffle the x, y and z
indices = np.arange(npoints)
np.random.seed(42)
np.random.shuffle(indices)
xs = x.take(indices)
ys = y.take(indices)
zs = z.take(indices)
df_feat = pd.DataFrame({'x': xs, 'y': ys})
df_target = pd.DataFrame({'z': zs})
df_nmi_target = nmi_target(df_feat=df_feat, df_target=df_target, n_neighbors=2)
assert df_nmi_target.shape == (2, 1)
assert df_nmi_target.loc['x']['z'] == pytest.approx(1.0)
assert df_nmi_target.loc['y']['z'] == pytest.approx(1.0)
# Test with one constant feature
c = np.ones(npoints) * 1.4
df_feat = pd.DataFrame({'x': x, 'y': y, 'c': c})
df_target = pd.DataFrame({'z': z})
df_nmi_target = nmi_target(df_feat=df_feat, df_target=df_target, n_neighbors=2)
assert df_nmi_target.shape == (2, 1)
assert df_nmi_target.loc['x']['z'] == pytest.approx(1.0)
assert df_nmi_target.loc['y']['z'] == pytest.approx(1.0)
df_nmi_target = nmi_target(df_feat=df_feat, df_target=df_target, drop_constant_features=False, n_neighbors=2)
assert df_nmi_target.shape == (3, 1)
assert df_nmi_target.loc['x']['z'] == pytest.approx(1.0)
assert df_nmi_target.loc['y']['z'] == pytest.approx(1.0)
assert df_nmi_target.loc['c']['z'] == pytest.approx(0.0)
# Test with unrelated data (grid)
x = np.linspace(start=2, stop=5, num=4)
z = np.linspace(start=3, stop=7, num=5)
x, z = np.meshgrid(x, z)
x = x.flatten()
z = z.flatten()
df_feat = pd.DataFrame({'x': x})
df_target = pd.DataFrame({'z': z})
df_nmi_target = nmi_target(df_feat=df_feat, df_target=df_target)
assert df_nmi_target.shape == (1, 1)
assert df_nmi_target.loc['x']['z'] == pytest.approx(0.0)
# Test initial checks
# Incompatible shapes
x = np.linspace(start=2, stop=3, num=5)
z = np.linspace(start=2, stop=3, num=8)
df_feat = pd.DataFrame({'x': x})
df_target = pd.DataFrame({'z': z})
with pytest.raises(ValueError, match=r'The input features DataFrame and the target variable DataFrame '
r'should contain the same number of data points.'):
nmi_target(df_feat=df_feat, df_target=df_target)
# Target DataFrame does not have exactly one column
x = np.linspace(start=2, stop=3, num=5)
z = np.linspace(start=2, stop=3, num=5)
df_feat = pd.DataFrame({'x': x})
df_target = pd.DataFrame({'z2': z, 'z': z})
with pytest.raises(ValueError, match=r'The target DataFrame should have exactly one column.'):
nmi_target(df_feat=df_feat, df_target=df_target)
# Test with some more real data (for which NMI is not just 0.0 or 1.0)
npoints = 200
np.random.seed(42)
x = np.random.rand(npoints)
z = 4 * x + 1.0 * np.random.rand(npoints)
df_feat = pd.DataFrame({'x': x})
df_target = pd.DataFrame({'z': z})
# Here we fix the random_state for the call to sklearn.feature_selection's mutual_info_regression so
# that we always get the same value.
df_nmi_target = nmi_target(df_feat=df_feat, df_target=df_target, random_state=42)
assert df_nmi_target.shape == (1, 1)
assert df_nmi_target.loc['x']['z'] == pytest.approx(0.3417665092162398)
def test_get_cross_nmi():
# Test with linear data (should get 1.0 mutual information, or very close due to algorithm used
# in mutual_info_regression)
npoints = 31
x = np.linspace(0.5, 3.5, npoints)
y = 2*x - 2
z = 4*x + 2
df_feat = pd.DataFrame({'x': x, 'y': y, 'z': z})
# Here we fix the number of neighbors for the call to sklearn.feature_selection's mutual_info_regression to 2 so
# that we get exactly 1 for the mutual information.
df_cross_nmi = get_cross_nmi(df_feat=df_feat, n_neighbors=2)
assert df_cross_nmi.shape == (3, 3)
for idx in df_cross_nmi.index:
for col in df_cross_nmi.columns:
assert df_cross_nmi.loc[idx][col] == pytest.approx(1.0)
# Same data shuffled
# Shuffle the x, y and z
indices = np.arange(npoints)
np.random.seed(42)
np.random.shuffle(indices)
xs = x.take(indices)
ys = y.take(indices)
zs = z.take(indices)
df_feat = pd.DataFrame({'x': xs, 'y': ys, 'z': zs})
df_cross_nmi = get_cross_nmi(df_feat=df_feat, n_neighbors=2)
assert df_cross_nmi.shape == (3, 3)
for idx in df_cross_nmi.index:
for col in df_cross_nmi.columns:
assert df_cross_nmi.loc[idx][col] == pytest.approx(1.0)
# Test with one constant feature
c = np.ones(npoints) * 1.4
df_feat = pd.DataFrame({'x': x, 'y': y, 'z': z, 'c': c})
df_cross_nmi = get_cross_nmi(df_feat=df_feat, n_neighbors=2)
assert df_cross_nmi.shape == (4, 4)
for idx in df_cross_nmi.index:
for col in df_cross_nmi.columns:
expected = 0.0 if idx == 'c' or col == 'c' else 1.0
assert df_cross_nmi.loc[idx][col] == pytest.approx(expected)
# Test with unrelated data (grid)
x = np.linspace(start=2, stop=5, num=4)
y = np.linspace(start=3, stop=7, num=5)
x, y = np.meshgrid(x, y)
x = x.flatten()
y = y.flatten()
df_feat = pd.DataFrame({'x': x, 'y': y})
df_cross_nmi = get_cross_nmi(df_feat=df_feat, n_neighbors=2)
assert df_cross_nmi.shape == (2, 2)
assert df_cross_nmi.loc['x']['y'] == pytest.approx(0.0)
assert df_cross_nmi.loc['y']['x'] == pytest.approx(0.0)
# Test with some more real data (for which NMI is not just 0.0 or 1.0)
npoints = 200
np.random.seed(42)
x = np.random.rand(npoints)
y = 4 * x + 1.0 * np.random.rand(npoints)
df_feat = | pd.DataFrame({'x': x, 'y': y}) | pandas.DataFrame |
import datetime
from datetime import timedelta
from distutils.version import LooseVersion
from io import BytesIO
import os
import re
from warnings import catch_warnings, simplefilter
import numpy as np
import pytest
from pandas.compat import is_platform_little_endian, is_platform_windows
import pandas.util._test_decorators as td
from pandas.core.dtypes.common import is_categorical_dtype
import pandas as pd
from pandas import (
Categorical,
CategoricalIndex,
DataFrame,
DatetimeIndex,
Index,
Int64Index,
MultiIndex,
RangeIndex,
Series,
Timestamp,
bdate_range,
concat,
date_range,
isna,
timedelta_range,
)
from pandas.tests.io.pytables.common import (
_maybe_remove,
create_tempfile,
ensure_clean_path,
ensure_clean_store,
safe_close,
safe_remove,
tables,
)
import pandas.util.testing as tm
from pandas.io.pytables import (
ClosedFileError,
HDFStore,
PossibleDataLossError,
Term,
read_hdf,
)
from pandas.io import pytables as pytables # noqa: E402 isort:skip
from pandas.io.pytables import TableIterator # noqa: E402 isort:skip
_default_compressor = "blosc"
ignore_natural_naming_warning = pytest.mark.filterwarnings(
"ignore:object name:tables.exceptions.NaturalNameWarning"
)
@pytest.mark.single
class TestHDFStore:
def test_format_kwarg_in_constructor(self, setup_path):
# GH 13291
with ensure_clean_path(setup_path) as path:
with pytest.raises(ValueError):
HDFStore(path, format="table")
def test_context(self, setup_path):
path = create_tempfile(setup_path)
try:
with HDFStore(path) as tbl:
raise ValueError("blah")
except ValueError:
pass
finally:
safe_remove(path)
try:
with HDFStore(path) as tbl:
tbl["a"] = tm.makeDataFrame()
with HDFStore(path) as tbl:
assert len(tbl) == 1
assert type(tbl["a"]) == DataFrame
finally:
safe_remove(path)
def test_conv_read_write(self, setup_path):
path = create_tempfile(setup_path)
try:
def roundtrip(key, obj, **kwargs):
obj.to_hdf(path, key, **kwargs)
return read_hdf(path, key)
o = tm.makeTimeSeries()
tm.assert_series_equal(o, roundtrip("series", o))
o = tm.makeStringSeries()
tm.assert_series_equal(o, roundtrip("string_series", o))
o = tm.makeDataFrame()
tm.assert_frame_equal(o, roundtrip("frame", o))
# table
df = DataFrame(dict(A=range(5), B=range(5)))
df.to_hdf(path, "table", append=True)
result = read_hdf(path, "table", where=["index>2"])
tm.assert_frame_equal(df[df.index > 2], result)
finally:
safe_remove(path)
def test_long_strings(self, setup_path):
# GH6166
df = DataFrame(
{"a": tm.rands_array(100, size=10)}, index=tm.rands_array(100, size=10)
)
with ensure_clean_store(setup_path) as store:
store.append("df", df, data_columns=["a"])
result = store.select("df")
tm.assert_frame_equal(df, result)
def test_api(self, setup_path):
# GH4584
# API issue when to_hdf doesn't accept append AND format args
with ensure_clean_path(setup_path) as path:
df = tm.makeDataFrame()
df.iloc[:10].to_hdf(path, "df", append=True, format="table")
df.iloc[10:].to_hdf(path, "df", append=True, format="table")
tm.assert_frame_equal(read_hdf(path, "df"), df)
# append to False
df.iloc[:10].to_hdf(path, "df", append=False, format="table")
df.iloc[10:].to_hdf(path, "df", append=True, format="table")
tm.assert_frame_equal(read_hdf(path, "df"), df)
with ensure_clean_path(setup_path) as path:
df = tm.makeDataFrame()
df.iloc[:10].to_hdf(path, "df", append=True)
df.iloc[10:].to_hdf(path, "df", append=True, format="table")
tm.assert_frame_equal(read_hdf(path, "df"), df)
# append to False
df.iloc[:10].to_hdf(path, "df", append=False, format="table")
df.iloc[10:].to_hdf(path, "df", append=True)
tm.assert_frame_equal(read_hdf(path, "df"), df)
with ensure_clean_path(setup_path) as path:
df = tm.makeDataFrame()
df.to_hdf(path, "df", append=False, format="fixed")
tm.assert_frame_equal(read_hdf(path, "df"), df)
df.to_hdf(path, "df", append=False, format="f")
tm.assert_frame_equal(read_hdf(path, "df"), df)
df.to_hdf(path, "df", append=False)
tm.assert_frame_equal(read_hdf(path, "df"), df)
df.to_hdf(path, "df")
tm.assert_frame_equal(read_hdf(path, "df"), df)
with ensure_clean_store(setup_path) as store:
path = store._path
df = tm.makeDataFrame()
_maybe_remove(store, "df")
store.append("df", df.iloc[:10], append=True, format="table")
store.append("df", df.iloc[10:], append=True, format="table")
tm.assert_frame_equal(store.select("df"), df)
# append to False
_maybe_remove(store, "df")
store.append("df", df.iloc[:10], append=False, format="table")
store.append("df", df.iloc[10:], append=True, format="table")
tm.assert_frame_equal(store.select("df"), df)
# formats
_maybe_remove(store, "df")
store.append("df", df.iloc[:10], append=False, format="table")
store.append("df", df.iloc[10:], append=True, format="table")
tm.assert_frame_equal(store.select("df"), df)
_maybe_remove(store, "df")
store.append("df", df.iloc[:10], append=False, format="table")
store.append("df", df.iloc[10:], append=True, format=None)
tm.assert_frame_equal(store.select("df"), df)
with ensure_clean_path(setup_path) as path:
# Invalid.
df = tm.makeDataFrame()
with pytest.raises(ValueError):
df.to_hdf(path, "df", append=True, format="f")
with pytest.raises(ValueError):
df.to_hdf(path, "df", append=True, format="fixed")
with pytest.raises(TypeError):
df.to_hdf(path, "df", append=True, format="foo")
with pytest.raises(TypeError):
df.to_hdf(path, "df", append=False, format="bar")
# File path doesn't exist
path = ""
with pytest.raises(FileNotFoundError):
read_hdf(path, "df")
def test_api_default_format(self, setup_path):
# default_format option
with ensure_clean_store(setup_path) as store:
df = tm.makeDataFrame()
pd.set_option("io.hdf.default_format", "fixed")
_maybe_remove(store, "df")
store.put("df", df)
assert not store.get_storer("df").is_table
with pytest.raises(ValueError):
store.append("df2", df)
pd.set_option("io.hdf.default_format", "table")
_maybe_remove(store, "df")
store.put("df", df)
assert store.get_storer("df").is_table
_maybe_remove(store, "df2")
store.append("df2", df)
assert store.get_storer("df").is_table
pd.set_option("io.hdf.default_format", None)
with ensure_clean_path(setup_path) as path:
df = tm.makeDataFrame()
pd.set_option("io.hdf.default_format", "fixed")
df.to_hdf(path, "df")
with HDFStore(path) as store:
assert not store.get_storer("df").is_table
with pytest.raises(ValueError):
df.to_hdf(path, "df2", append=True)
pd.set_option("io.hdf.default_format", "table")
df.to_hdf(path, "df3")
with HDFStore(path) as store:
assert store.get_storer("df3").is_table
df.to_hdf(path, "df4", append=True)
with HDFStore(path) as store:
assert store.get_storer("df4").is_table
pd.set_option("io.hdf.default_format", None)
def test_keys(self, setup_path):
with ensure_clean_store(setup_path) as store:
store["a"] = tm.makeTimeSeries()
store["b"] = tm.makeStringSeries()
store["c"] = tm.makeDataFrame()
assert len(store) == 3
expected = {"/a", "/b", "/c"}
assert set(store.keys()) == expected
assert set(store) == expected
def test_keys_ignore_hdf_softlink(self, setup_path):
# GH 20523
# Puts a softlink into HDF file and rereads
with ensure_clean_store(setup_path) as store:
df = DataFrame(dict(A=range(5), B=range(5)))
store.put("df", df)
assert store.keys() == ["/df"]
store._handle.create_soft_link(store._handle.root, "symlink", "df")
# Should ignore the softlink
assert store.keys() == ["/df"]
def test_iter_empty(self, setup_path):
with ensure_clean_store(setup_path) as store:
# GH 12221
assert list(store) == []
def test_repr(self, setup_path):
with ensure_clean_store(setup_path) as store:
repr(store)
store.info()
store["a"] = tm.makeTimeSeries()
store["b"] = tm.makeStringSeries()
store["c"] = tm.makeDataFrame()
df = tm.makeDataFrame()
df["obj1"] = "foo"
df["obj2"] = "bar"
df["bool1"] = df["A"] > 0
df["bool2"] = df["B"] > 0
df["bool3"] = True
df["int1"] = 1
df["int2"] = 2
df["timestamp1"] = Timestamp("20010102")
df["timestamp2"] = Timestamp("20010103")
df["datetime1"] = datetime.datetime(2001, 1, 2, 0, 0)
df["datetime2"] = datetime.datetime(2001, 1, 3, 0, 0)
df.loc[3:6, ["obj1"]] = np.nan
df = df._consolidate()._convert(datetime=True)
with catch_warnings(record=True):
simplefilter("ignore", pd.errors.PerformanceWarning)
store["df"] = df
# make a random group in hdf space
store._handle.create_group(store._handle.root, "bah")
assert store.filename in repr(store)
assert store.filename in str(store)
store.info()
# storers
with ensure_clean_store(setup_path) as store:
df = tm.makeDataFrame()
store.append("df", df)
s = store.get_storer("df")
repr(s)
str(s)
@ignore_natural_naming_warning
def test_contains(self, setup_path):
with ensure_clean_store(setup_path) as store:
store["a"] = tm.makeTimeSeries()
store["b"] = tm.makeDataFrame()
store["foo/bar"] = tm.makeDataFrame()
assert "a" in store
assert "b" in store
assert "c" not in store
assert "foo/bar" in store
assert "/foo/bar" in store
assert "/foo/b" not in store
assert "bar" not in store
# gh-2694: tables.NaturalNameWarning
with catch_warnings(record=True):
store["node())"] = tm.makeDataFrame()
assert "node())" in store
def test_versioning(self, setup_path):
with ensure_clean_store(setup_path) as store:
store["a"] = tm.makeTimeSeries()
store["b"] = tm.makeDataFrame()
df = tm.makeTimeDataFrame()
_maybe_remove(store, "df1")
store.append("df1", df[:10])
store.append("df1", df[10:])
assert store.root.a._v_attrs.pandas_version == "0.15.2"
assert store.root.b._v_attrs.pandas_version == "0.15.2"
assert store.root.df1._v_attrs.pandas_version == "0.15.2"
# write a file and wipe its versioning
_maybe_remove(store, "df2")
store.append("df2", df)
# this is an error because its table_type is appendable, but no
# version info
store.get_node("df2")._v_attrs.pandas_version = None
with pytest.raises(Exception):
store.select("df2")
def test_mode(self, setup_path):
df = tm.makeTimeDataFrame()
def check(mode):
with ensure_clean_path(setup_path) as path:
# constructor
if mode in ["r", "r+"]:
with pytest.raises(IOError):
HDFStore(path, mode=mode)
else:
store = HDFStore(path, mode=mode)
assert store._handle.mode == mode
store.close()
with ensure_clean_path(setup_path) as path:
# context
if mode in ["r", "r+"]:
with pytest.raises(IOError):
with HDFStore(path, mode=mode) as store: # noqa
pass
else:
with HDFStore(path, mode=mode) as store:
assert store._handle.mode == mode
with ensure_clean_path(setup_path) as path:
# conv write
if mode in ["r", "r+"]:
with pytest.raises(IOError):
df.to_hdf(path, "df", mode=mode)
df.to_hdf(path, "df", mode="w")
else:
df.to_hdf(path, "df", mode=mode)
# conv read
if mode in ["w"]:
with pytest.raises(ValueError):
read_hdf(path, "df", mode=mode)
else:
result = read_hdf(path, "df", mode=mode)
tm.assert_frame_equal(result, df)
def check_default_mode():
# read_hdf uses default mode
with ensure_clean_path(setup_path) as path:
df.to_hdf(path, "df", mode="w")
result = read_hdf(path, "df")
tm.assert_frame_equal(result, df)
check("r")
check("r+")
check("a")
check("w")
check_default_mode()
def test_reopen_handle(self, setup_path):
with ensure_clean_path(setup_path) as path:
store = HDFStore(path, mode="a")
store["a"] = tm.makeTimeSeries()
# invalid mode change
with pytest.raises(PossibleDataLossError):
store.open("w")
store.close()
assert not store.is_open
# truncation ok here
store.open("w")
assert store.is_open
assert len(store) == 0
store.close()
assert not store.is_open
store = HDFStore(path, mode="a")
store["a"] = tm.makeTimeSeries()
# reopen as read
store.open("r")
assert store.is_open
assert len(store) == 1
assert store._mode == "r"
store.close()
assert not store.is_open
# reopen as append
store.open("a")
assert store.is_open
assert len(store) == 1
assert store._mode == "a"
store.close()
assert not store.is_open
# reopen as append (again)
store.open("a")
assert store.is_open
assert len(store) == 1
assert store._mode == "a"
store.close()
assert not store.is_open
def test_open_args(self, setup_path):
with ensure_clean_path(setup_path) as path:
df = tm.makeDataFrame()
# create an in memory store
store = HDFStore(
path, mode="a", driver="H5FD_CORE", driver_core_backing_store=0
)
store["df"] = df
store.append("df2", df)
tm.assert_frame_equal(store["df"], df)
tm.assert_frame_equal(store["df2"], df)
store.close()
# the file should not have actually been written
assert not os.path.exists(path)
def test_flush(self, setup_path):
with ensure_clean_store(setup_path) as store:
store["a"] = tm.makeTimeSeries()
store.flush()
store.flush(fsync=True)
def test_get(self, setup_path):
with ensure_clean_store(setup_path) as store:
store["a"] = tm.makeTimeSeries()
left = store.get("a")
right = store["a"]
tm.assert_series_equal(left, right)
left = store.get("/a")
right = store["/a"]
tm.assert_series_equal(left, right)
with pytest.raises(KeyError, match="'No object named b in the file'"):
store.get("b")
@pytest.mark.parametrize(
"where, expected",
[
(
"/",
{
"": ({"first_group", "second_group"}, set()),
"/first_group": (set(), {"df1", "df2"}),
"/second_group": ({"third_group"}, {"df3", "s1"}),
"/second_group/third_group": (set(), {"df4"}),
},
),
(
"/second_group",
{
"/second_group": ({"third_group"}, {"df3", "s1"}),
"/second_group/third_group": (set(), {"df4"}),
},
),
],
)
def test_walk(self, where, expected, setup_path):
# GH10143
objs = {
"df1": pd.DataFrame([1, 2, 3]),
"df2": pd.DataFrame([4, 5, 6]),
"df3": pd.DataFrame([6, 7, 8]),
"df4": pd.DataFrame([9, 10, 11]),
"s1": pd.Series([10, 9, 8]),
# Next 3 items aren't pandas objects and should be ignored
"a1": np.array([[1, 2, 3], [4, 5, 6]]),
"tb1": np.array([(1, 2, 3), (4, 5, 6)], dtype="i,i,i"),
"tb2": np.array([(7, 8, 9), (10, 11, 12)], dtype="i,i,i"),
}
with ensure_clean_store("walk_groups.hdf", mode="w") as store:
store.put("/first_group/df1", objs["df1"])
store.put("/first_group/df2", objs["df2"])
store.put("/second_group/df3", objs["df3"])
store.put("/second_group/s1", objs["s1"])
store.put("/second_group/third_group/df4", objs["df4"])
# Create non-pandas objects
store._handle.create_array("/first_group", "a1", objs["a1"])
store._handle.create_table("/first_group", "tb1", obj=objs["tb1"])
store._handle.create_table("/second_group", "tb2", obj=objs["tb2"])
assert len(list(store.walk(where=where))) == len(expected)
for path, groups, leaves in store.walk(where=where):
assert path in expected
expected_groups, expected_frames = expected[path]
assert expected_groups == set(groups)
assert expected_frames == set(leaves)
for leaf in leaves:
frame_path = "/".join([path, leaf])
obj = store.get(frame_path)
if "df" in leaf:
tm.assert_frame_equal(obj, objs[leaf])
else:
tm.assert_series_equal(obj, objs[leaf])
def test_getattr(self, setup_path):
with ensure_clean_store(setup_path) as store:
s = tm.makeTimeSeries()
store["a"] = s
# test attribute access
result = store.a
tm.assert_series_equal(result, s)
result = getattr(store, "a")
tm.assert_series_equal(result, s)
df = tm.makeTimeDataFrame()
store["df"] = df
result = store.df
tm.assert_frame_equal(result, df)
# errors
for x in ["d", "mode", "path", "handle", "complib"]:
with pytest.raises(AttributeError):
getattr(store, x)
# not stores
for x in ["mode", "path", "handle", "complib"]:
getattr(store, "_{x}".format(x=x))
def test_put(self, setup_path):
with ensure_clean_store(setup_path) as store:
ts = tm.makeTimeSeries()
df = tm.makeTimeDataFrame()
store["a"] = ts
store["b"] = df[:10]
store["foo/bar/bah"] = df[:10]
store["foo"] = df[:10]
store["/foo"] = df[:10]
store.put("c", df[:10], format="table")
# not OK, not a table
with pytest.raises(ValueError):
store.put("b", df[10:], append=True)
# node does not currently exist, test _is_table_type returns False
# in this case
_maybe_remove(store, "f")
with pytest.raises(ValueError):
store.put("f", df[10:], append=True)
# can't put to a table (use append instead)
with pytest.raises(ValueError):
store.put("c", df[10:], append=True)
# overwrite table
store.put("c", df[:10], format="table", append=False)
tm.assert_frame_equal(df[:10], store["c"])
def test_put_string_index(self, setup_path):
with ensure_clean_store(setup_path) as store:
index = Index(
["I am a very long string index: {i}".format(i=i) for i in range(20)]
)
s = Series(np.arange(20), index=index)
df = DataFrame({"A": s, "B": s})
store["a"] = s
tm.assert_series_equal(store["a"], s)
store["b"] = df
tm.assert_frame_equal(store["b"], df)
# mixed length
index = Index(
["abcdefghijklmnopqrstuvwxyz1234567890"]
+ ["I am a very long string index: {i}".format(i=i) for i in range(20)]
)
s = Series(np.arange(21), index=index)
df = DataFrame({"A": s, "B": s})
store["a"] = s
tm.assert_series_equal(store["a"], s)
store["b"] = df
tm.assert_frame_equal(store["b"], df)
def test_put_compression(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = tm.makeTimeDataFrame()
store.put("c", df, format="table", complib="zlib")
tm.assert_frame_equal(store["c"], df)
# can't compress if format='fixed'
with pytest.raises(ValueError):
store.put("b", df, format="fixed", complib="zlib")
@td.skip_if_windows_python_3
def test_put_compression_blosc(self, setup_path):
df = tm.makeTimeDataFrame()
with ensure_clean_store(setup_path) as store:
# can't compress if format='fixed'
with pytest.raises(ValueError):
store.put("b", df, format="fixed", complib="blosc")
store.put("c", df, format="table", complib="blosc")
tm.assert_frame_equal(store["c"], df)
def test_complibs_default_settings(self, setup_path):
# GH15943
df = tm.makeDataFrame()
# Set complevel and check if complib is automatically set to
# default value
with ensure_clean_path(setup_path) as tmpfile:
df.to_hdf(tmpfile, "df", complevel=9)
result = pd.read_hdf(tmpfile, "df")
tm.assert_frame_equal(result, df)
with tables.open_file(tmpfile, mode="r") as h5file:
for node in h5file.walk_nodes(where="/df", classname="Leaf"):
assert node.filters.complevel == 9
assert node.filters.complib == "zlib"
# Set complib and check to see if compression is disabled
with ensure_clean_path(setup_path) as tmpfile:
df.to_hdf(tmpfile, "df", complib="zlib")
result = pd.read_hdf(tmpfile, "df")
tm.assert_frame_equal(result, df)
with tables.open_file(tmpfile, mode="r") as h5file:
for node in h5file.walk_nodes(where="/df", classname="Leaf"):
assert node.filters.complevel == 0
assert node.filters.complib is None
# Check if not setting complib or complevel results in no compression
with ensure_clean_path(setup_path) as tmpfile:
df.to_hdf(tmpfile, "df")
result = pd.read_hdf(tmpfile, "df")
tm.assert_frame_equal(result, df)
with tables.open_file(tmpfile, mode="r") as h5file:
for node in h5file.walk_nodes(where="/df", classname="Leaf"):
assert node.filters.complevel == 0
assert node.filters.complib is None
# Check if file-defaults can be overridden on a per table basis
with ensure_clean_path(setup_path) as tmpfile:
store = pd.HDFStore(tmpfile)
store.append("dfc", df, complevel=9, complib="blosc")
store.append("df", df)
store.close()
with tables.open_file(tmpfile, mode="r") as h5file:
for node in h5file.walk_nodes(where="/df", classname="Leaf"):
assert node.filters.complevel == 0
assert node.filters.complib is None
for node in h5file.walk_nodes(where="/dfc", classname="Leaf"):
assert node.filters.complevel == 9
assert node.filters.complib == "blosc"
def test_complibs(self, setup_path):
# GH14478
df = tm.makeDataFrame()
# Building list of all complibs and complevels tuples
all_complibs = tables.filters.all_complibs
# Remove lzo if its not available on this platform
if not tables.which_lib_version("lzo"):
all_complibs.remove("lzo")
# Remove bzip2 if its not available on this platform
if not tables.which_lib_version("bzip2"):
all_complibs.remove("bzip2")
all_levels = range(0, 10)
all_tests = [(lib, lvl) for lib in all_complibs for lvl in all_levels]
for (lib, lvl) in all_tests:
with ensure_clean_path(setup_path) as tmpfile:
gname = "foo"
# Write and read file to see if data is consistent
df.to_hdf(tmpfile, gname, complib=lib, complevel=lvl)
result = pd.read_hdf(tmpfile, gname)
tm.assert_frame_equal(result, df)
# Open file and check metadata
# for correct amount of compression
h5table = tables.open_file(tmpfile, mode="r")
for node in h5table.walk_nodes(where="/" + gname, classname="Leaf"):
assert node.filters.complevel == lvl
if lvl == 0:
assert node.filters.complib is None
else:
assert node.filters.complib == lib
h5table.close()
def test_put_integer(self, setup_path):
# non-date, non-string index
df = DataFrame(np.random.randn(50, 100))
self._check_roundtrip(df, tm.assert_frame_equal, setup_path)
@td.xfail_non_writeable
def test_put_mixed_type(self, setup_path):
df = tm.makeTimeDataFrame()
df["obj1"] = "foo"
df["obj2"] = "bar"
df["bool1"] = df["A"] > 0
df["bool2"] = df["B"] > 0
df["bool3"] = True
df["int1"] = 1
df["int2"] = 2
df["timestamp1"] = Timestamp("20010102")
df["timestamp2"] = Timestamp("20010103")
df["datetime1"] = datetime.datetime(2001, 1, 2, 0, 0)
df["datetime2"] = datetime.datetime(2001, 1, 3, 0, 0)
df.loc[3:6, ["obj1"]] = np.nan
df = df._consolidate()._convert(datetime=True)
with ensure_clean_store(setup_path) as store:
_maybe_remove(store, "df")
# PerformanceWarning
with catch_warnings(record=True):
simplefilter("ignore", pd.errors.PerformanceWarning)
store.put("df", df)
expected = store.get("df")
tm.assert_frame_equal(expected, df)
@pytest.mark.filterwarnings(
"ignore:object name:tables.exceptions.NaturalNameWarning"
)
def test_append(self, setup_path):
with ensure_clean_store(setup_path) as store:
# this is allowed by almost always don't want to do it
# tables.NaturalNameWarning):
with catch_warnings(record=True):
df = tm.makeTimeDataFrame()
_maybe_remove(store, "df1")
store.append("df1", df[:10])
store.append("df1", df[10:])
tm.assert_frame_equal(store["df1"], df)
_maybe_remove(store, "df2")
store.put("df2", df[:10], format="table")
store.append("df2", df[10:])
tm.assert_frame_equal(store["df2"], df)
_maybe_remove(store, "df3")
store.append("/df3", df[:10])
store.append("/df3", df[10:])
tm.assert_frame_equal(store["df3"], df)
# this is allowed by almost always don't want to do it
# tables.NaturalNameWarning
_maybe_remove(store, "/df3 foo")
store.append("/df3 foo", df[:10])
store.append("/df3 foo", df[10:])
tm.assert_frame_equal(store["df3 foo"], df)
# dtype issues - mizxed type in a single object column
df = DataFrame(data=[[1, 2], [0, 1], [1, 2], [0, 0]])
df["mixed_column"] = "testing"
df.loc[2, "mixed_column"] = np.nan
_maybe_remove(store, "df")
store.append("df", df)
tm.assert_frame_equal(store["df"], df)
# uints - test storage of uints
uint_data = DataFrame(
{
"u08": Series(
np.random.randint(0, high=255, size=5), dtype=np.uint8
),
"u16": Series(
np.random.randint(0, high=65535, size=5), dtype=np.uint16
),
"u32": Series(
np.random.randint(0, high=2 ** 30, size=5), dtype=np.uint32
),
"u64": Series(
[2 ** 58, 2 ** 59, 2 ** 60, 2 ** 61, 2 ** 62],
dtype=np.uint64,
),
},
index=np.arange(5),
)
_maybe_remove(store, "uints")
store.append("uints", uint_data)
tm.assert_frame_equal(store["uints"], uint_data)
# uints - test storage of uints in indexable columns
_maybe_remove(store, "uints")
# 64-bit indices not yet supported
store.append("uints", uint_data, data_columns=["u08", "u16", "u32"])
tm.assert_frame_equal(store["uints"], uint_data)
def test_append_series(self, setup_path):
with ensure_clean_store(setup_path) as store:
# basic
ss = tm.makeStringSeries()
ts = tm.makeTimeSeries()
ns = Series(np.arange(100))
store.append("ss", ss)
result = store["ss"]
tm.assert_series_equal(result, ss)
assert result.name is None
store.append("ts", ts)
result = store["ts"]
tm.assert_series_equal(result, ts)
assert result.name is None
ns.name = "foo"
store.append("ns", ns)
result = store["ns"]
tm.assert_series_equal(result, ns)
assert result.name == ns.name
# select on the values
expected = ns[ns > 60]
result = store.select("ns", "foo>60")
tm.assert_series_equal(result, expected)
# select on the index and values
expected = ns[(ns > 70) & (ns.index < 90)]
result = store.select("ns", "foo>70 and index<90")
tm.assert_series_equal(result, expected)
# multi-index
mi = DataFrame(np.random.randn(5, 1), columns=["A"])
mi["B"] = np.arange(len(mi))
mi["C"] = "foo"
mi.loc[3:5, "C"] = "bar"
mi.set_index(["C", "B"], inplace=True)
s = mi.stack()
s.index = s.index.droplevel(2)
store.append("mi", s)
tm.assert_series_equal(store["mi"], s)
def test_store_index_types(self, setup_path):
# GH5386
# test storing various index types
with ensure_clean_store(setup_path) as store:
def check(format, index):
df = DataFrame(np.random.randn(10, 2), columns=list("AB"))
df.index = index(len(df))
_maybe_remove(store, "df")
store.put("df", df, format=format)
tm.assert_frame_equal(df, store["df"])
for index in [
tm.makeFloatIndex,
tm.makeStringIndex,
tm.makeIntIndex,
tm.makeDateIndex,
]:
check("table", index)
check("fixed", index)
# period index currently broken for table
# seee GH7796 FIXME
check("fixed", tm.makePeriodIndex)
# check('table',tm.makePeriodIndex)
# unicode
index = tm.makeUnicodeIndex
check("table", index)
check("fixed", index)
@pytest.mark.skipif(
not is_platform_little_endian(), reason="reason platform is not little endian"
)
def test_encoding(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = DataFrame(dict(A="foo", B="bar"), index=range(5))
df.loc[2, "A"] = np.nan
df.loc[3, "B"] = np.nan
_maybe_remove(store, "df")
store.append("df", df, encoding="ascii")
tm.assert_frame_equal(store["df"], df)
expected = df.reindex(columns=["A"])
result = store.select("df", Term("columns=A", encoding="ascii"))
tm.assert_frame_equal(result, expected)
@pytest.mark.parametrize(
"val",
[
[b"E\xc9, 17", b"", b"a", b"b", b"c"],
[b"E\xc9, 17", b"a", b"b", b"c"],
[b"EE, 17", b"", b"a", b"b", b"c"],
[b"E\xc9, 17", b"\xf8\xfc", b"a", b"b", b"c"],
[b"", b"a", b"b", b"c"],
[b"\xf8\xfc", b"a", b"b", b"c"],
[b"A\xf8\xfc", b"", b"a", b"b", b"c"],
[np.nan, b"", b"b", b"c"],
[b"A\xf8\xfc", np.nan, b"", b"b", b"c"],
],
)
@pytest.mark.parametrize("dtype", ["category", object])
def test_latin_encoding(self, setup_path, dtype, val):
enc = "latin-1"
nan_rep = ""
key = "data"
val = [x.decode(enc) if isinstance(x, bytes) else x for x in val]
ser = pd.Series(val, dtype=dtype)
with ensure_clean_path(setup_path) as store:
ser.to_hdf(store, key, format="table", encoding=enc, nan_rep=nan_rep)
retr = read_hdf(store, key)
s_nan = ser.replace(nan_rep, np.nan)
if is_categorical_dtype(s_nan):
assert is_categorical_dtype(retr)
tm.assert_series_equal(
s_nan, retr, check_dtype=False, check_categorical=False
)
else:
tm.assert_series_equal(s_nan, retr)
# FIXME: don't leave commented-out
# fails:
# for x in examples:
# roundtrip(s, nan_rep=b'\xf8\xfc')
def test_append_some_nans(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = DataFrame(
{
"A": Series(np.random.randn(20)).astype("int32"),
"A1": np.random.randn(20),
"A2": np.random.randn(20),
"B": "foo",
"C": "bar",
"D": Timestamp("20010101"),
"E": datetime.datetime(2001, 1, 2, 0, 0),
},
index=np.arange(20),
)
# some nans
_maybe_remove(store, "df1")
df.loc[0:15, ["A1", "B", "D", "E"]] = np.nan
store.append("df1", df[:10])
store.append("df1", df[10:])
tm.assert_frame_equal(store["df1"], df)
# first column
df1 = df.copy()
df1.loc[:, "A1"] = np.nan
_maybe_remove(store, "df1")
store.append("df1", df1[:10])
store.append("df1", df1[10:])
tm.assert_frame_equal(store["df1"], df1)
# 2nd column
df2 = df.copy()
df2.loc[:, "A2"] = np.nan
_maybe_remove(store, "df2")
store.append("df2", df2[:10])
store.append("df2", df2[10:])
tm.assert_frame_equal(store["df2"], df2)
# datetimes
df3 = df.copy()
df3.loc[:, "E"] = np.nan
_maybe_remove(store, "df3")
store.append("df3", df3[:10])
store.append("df3", df3[10:])
tm.assert_frame_equal(store["df3"], df3)
def test_append_all_nans(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = DataFrame(
{"A1": np.random.randn(20), "A2": np.random.randn(20)},
index=np.arange(20),
)
df.loc[0:15, :] = np.nan
# nan some entire rows (dropna=True)
_maybe_remove(store, "df")
store.append("df", df[:10], dropna=True)
store.append("df", df[10:], dropna=True)
tm.assert_frame_equal(store["df"], df[-4:])
# nan some entire rows (dropna=False)
_maybe_remove(store, "df2")
store.append("df2", df[:10], dropna=False)
store.append("df2", df[10:], dropna=False)
tm.assert_frame_equal(store["df2"], df)
# tests the option io.hdf.dropna_table
pd.set_option("io.hdf.dropna_table", False)
_maybe_remove(store, "df3")
store.append("df3", df[:10])
store.append("df3", df[10:])
tm.assert_frame_equal(store["df3"], df)
pd.set_option("io.hdf.dropna_table", True)
_maybe_remove(store, "df4")
store.append("df4", df[:10])
store.append("df4", df[10:])
tm.assert_frame_equal(store["df4"], df[-4:])
# nan some entire rows (string are still written!)
df = DataFrame(
{
"A1": np.random.randn(20),
"A2": np.random.randn(20),
"B": "foo",
"C": "bar",
},
index=np.arange(20),
)
df.loc[0:15, :] = np.nan
_maybe_remove(store, "df")
store.append("df", df[:10], dropna=True)
store.append("df", df[10:], dropna=True)
tm.assert_frame_equal(store["df"], df)
_maybe_remove(store, "df2")
store.append("df2", df[:10], dropna=False)
store.append("df2", df[10:], dropna=False)
tm.assert_frame_equal(store["df2"], df)
# nan some entire rows (but since we have dates they are still
# written!)
df = DataFrame(
{
"A1": np.random.randn(20),
"A2": np.random.randn(20),
"B": "foo",
"C": "bar",
"D": Timestamp("20010101"),
"E": datetime.datetime(2001, 1, 2, 0, 0),
},
index=np.arange(20),
)
df.loc[0:15, :] = np.nan
_maybe_remove(store, "df")
store.append("df", df[:10], dropna=True)
store.append("df", df[10:], dropna=True)
tm.assert_frame_equal(store["df"], df)
_maybe_remove(store, "df2")
store.append("df2", df[:10], dropna=False)
store.append("df2", df[10:], dropna=False)
tm.assert_frame_equal(store["df2"], df)
# Test to make sure defaults are to not drop.
# Corresponding to Issue 9382
df_with_missing = DataFrame(
{"col1": [0, np.nan, 2], "col2": [1, np.nan, np.nan]}
)
with ensure_clean_path(setup_path) as path:
df_with_missing.to_hdf(path, "df_with_missing", format="table")
reloaded = read_hdf(path, "df_with_missing")
tm.assert_frame_equal(df_with_missing, reloaded)
def test_read_missing_key_close_store(self, setup_path):
# GH 25766
with ensure_clean_path(setup_path) as path:
df = pd.DataFrame({"a": range(2), "b": range(2)})
df.to_hdf(path, "k1")
with pytest.raises(KeyError, match="'No object named k2 in the file'"):
pd.read_hdf(path, "k2")
# smoke test to test that file is properly closed after
# read with KeyError before another write
df.to_hdf(path, "k2")
def test_read_missing_key_opened_store(self, setup_path):
# GH 28699
with ensure_clean_path(setup_path) as path:
df = pd.DataFrame({"a": range(2), "b": range(2)})
df.to_hdf(path, "k1")
store = pd.HDFStore(path, "r")
with pytest.raises(KeyError, match="'No object named k2 in the file'"):
pd.read_hdf(store, "k2")
# Test that the file is still open after a KeyError and that we can
# still read from it.
pd.read_hdf(store, "k1")
def test_append_frame_column_oriented(self, setup_path):
with ensure_clean_store(setup_path) as store:
# column oriented
df = tm.makeTimeDataFrame()
_maybe_remove(store, "df1")
store.append("df1", df.iloc[:, :2], axes=["columns"])
store.append("df1", df.iloc[:, 2:])
tm.assert_frame_equal(store["df1"], df)
result = store.select("df1", "columns=A")
expected = df.reindex(columns=["A"])
tm.assert_frame_equal(expected, result)
# selection on the non-indexable
result = store.select("df1", ("columns=A", "index=df.index[0:4]"))
expected = df.reindex(columns=["A"], index=df.index[0:4])
tm.assert_frame_equal(expected, result)
# this isn't supported
with pytest.raises(TypeError):
store.select("df1", "columns=A and index>df.index[4]")
def test_append_with_different_block_ordering(self, setup_path):
# GH 4096; using same frames, but different block orderings
with ensure_clean_store(setup_path) as store:
for i in range(10):
df = DataFrame(np.random.randn(10, 2), columns=list("AB"))
df["index"] = range(10)
df["index"] += i * 10
df["int64"] = Series([1] * len(df), dtype="int64")
df["int16"] = Series([1] * len(df), dtype="int16")
if i % 2 == 0:
del df["int64"]
df["int64"] = Series([1] * len(df), dtype="int64")
if i % 3 == 0:
a = df.pop("A")
df["A"] = a
df.set_index("index", inplace=True)
store.append("df", df)
# test a different ordering but with more fields (like invalid
# combinate)
with ensure_clean_store(setup_path) as store:
df = DataFrame(np.random.randn(10, 2), columns=list("AB"), dtype="float64")
df["int64"] = Series([1] * len(df), dtype="int64")
df["int16"] = Series([1] * len(df), dtype="int16")
store.append("df", df)
# store additional fields in different blocks
df["int16_2"] = Series([1] * len(df), dtype="int16")
with pytest.raises(ValueError):
store.append("df", df)
# store multile additional fields in different blocks
df["float_3"] = Series([1.0] * len(df), dtype="float64")
with pytest.raises(ValueError):
store.append("df", df)
def test_append_with_strings(self, setup_path):
with ensure_clean_store(setup_path) as store:
with catch_warnings(record=True):
def check_col(key, name, size):
assert (
getattr(store.get_storer(key).table.description, name).itemsize
== size
)
# avoid truncation on elements
df = DataFrame([[123, "asdqwerty"], [345, "dggnhebbsdfbdfb"]])
store.append("df_big", df)
tm.assert_frame_equal(store.select("df_big"), df)
check_col("df_big", "values_block_1", 15)
# appending smaller string ok
df2 = DataFrame([[124, "asdqy"], [346, "dggnhefbdfb"]])
store.append("df_big", df2)
expected = concat([df, df2])
tm.assert_frame_equal(store.select("df_big"), expected)
check_col("df_big", "values_block_1", 15)
# avoid truncation on elements
df = DataFrame([[123, "asdqwerty"], [345, "dggnhebbsdfbdfb"]])
store.append("df_big2", df, min_itemsize={"values": 50})
tm.assert_frame_equal(store.select("df_big2"), df)
check_col("df_big2", "values_block_1", 50)
# bigger string on next append
store.append("df_new", df)
df_new = DataFrame(
[[124, "abcdefqhij"], [346, "abcdefghijklmnopqrtsuvwxyz"]]
)
with pytest.raises(ValueError):
store.append("df_new", df_new)
# min_itemsize on Series index (GH 11412)
df = tm.makeMixedDataFrame().set_index("C")
store.append("ss", df["B"], min_itemsize={"index": 4})
tm.assert_series_equal(store.select("ss"), df["B"])
# same as above, with data_columns=True
store.append(
"ss2", df["B"], data_columns=True, min_itemsize={"index": 4}
)
tm.assert_series_equal(store.select("ss2"), df["B"])
# min_itemsize in index without appending (GH 10381)
store.put("ss3", df, format="table", min_itemsize={"index": 6})
# just make sure there is a longer string:
df2 = df.copy().reset_index().assign(C="longer").set_index("C")
store.append("ss3", df2)
tm.assert_frame_equal(store.select("ss3"), pd.concat([df, df2]))
# same as above, with a Series
store.put("ss4", df["B"], format="table", min_itemsize={"index": 6})
store.append("ss4", df2["B"])
tm.assert_series_equal(
store.select("ss4"), pd.concat([df["B"], df2["B"]])
)
# with nans
_maybe_remove(store, "df")
df = tm.makeTimeDataFrame()
df["string"] = "foo"
df.loc[1:4, "string"] = np.nan
df["string2"] = "bar"
df.loc[4:8, "string2"] = np.nan
df["string3"] = "bah"
df.loc[1:, "string3"] = np.nan
store.append("df", df)
result = store.select("df")
tm.assert_frame_equal(result, df)
with ensure_clean_store(setup_path) as store:
def check_col(key, name, size):
assert getattr(
store.get_storer(key).table.description, name
).itemsize, size
df = DataFrame(dict(A="foo", B="bar"), index=range(10))
# a min_itemsize that creates a data_column
_maybe_remove(store, "df")
store.append("df", df, min_itemsize={"A": 200})
check_col("df", "A", 200)
assert store.get_storer("df").data_columns == ["A"]
# a min_itemsize that creates a data_column2
_maybe_remove(store, "df")
store.append("df", df, data_columns=["B"], min_itemsize={"A": 200})
check_col("df", "A", 200)
assert store.get_storer("df").data_columns == ["B", "A"]
# a min_itemsize that creates a data_column2
_maybe_remove(store, "df")
store.append("df", df, data_columns=["B"], min_itemsize={"values": 200})
check_col("df", "B", 200)
check_col("df", "values_block_0", 200)
assert store.get_storer("df").data_columns == ["B"]
# infer the .typ on subsequent appends
_maybe_remove(store, "df")
store.append("df", df[:5], min_itemsize=200)
store.append("df", df[5:], min_itemsize=200)
tm.assert_frame_equal(store["df"], df)
# invalid min_itemsize keys
df = DataFrame(["foo", "foo", "foo", "barh", "barh", "barh"], columns=["A"])
_maybe_remove(store, "df")
with pytest.raises(ValueError):
store.append("df", df, min_itemsize={"foo": 20, "foobar": 20})
def test_append_with_empty_string(self, setup_path):
with ensure_clean_store(setup_path) as store:
# with all empty strings (GH 12242)
df = DataFrame({"x": ["a", "b", "c", "d", "e", "f", ""]})
store.append("df", df[:-1], min_itemsize={"x": 1})
store.append("df", df[-1:], min_itemsize={"x": 1})
tm.assert_frame_equal(store.select("df"), df)
def test_to_hdf_with_min_itemsize(self, setup_path):
with ensure_clean_path(setup_path) as path:
# min_itemsize in index with to_hdf (GH 10381)
df = tm.makeMixedDataFrame().set_index("C")
df.to_hdf(path, "ss3", format="table", min_itemsize={"index": 6})
# just make sure there is a longer string:
df2 = df.copy().reset_index().assign(C="longer").set_index("C")
df2.to_hdf(path, "ss3", append=True, format="table")
tm.assert_frame_equal(pd.read_hdf(path, "ss3"), pd.concat([df, df2]))
# same as above, with a Series
df["B"].to_hdf(path, "ss4", format="table", min_itemsize={"index": 6})
df2["B"].to_hdf(path, "ss4", append=True, format="table")
tm.assert_series_equal(
pd.read_hdf(path, "ss4"), pd.concat([df["B"], df2["B"]])
)
@pytest.mark.parametrize(
"format", [pytest.param("fixed", marks=td.xfail_non_writeable), "table"]
)
def test_to_hdf_errors(self, format, setup_path):
data = ["\ud800foo"]
ser = pd.Series(data, index=pd.Index(data))
with ensure_clean_path(setup_path) as path:
# GH 20835
ser.to_hdf(path, "table", format=format, errors="surrogatepass")
result = pd.read_hdf(path, "table", errors="surrogatepass")
tm.assert_series_equal(result, ser)
def test_append_with_data_columns(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = tm.makeTimeDataFrame()
df.iloc[0, df.columns.get_loc("B")] = 1.0
_maybe_remove(store, "df")
store.append("df", df[:2], data_columns=["B"])
store.append("df", df[2:])
tm.assert_frame_equal(store["df"], df)
# check that we have indices created
assert store._handle.root.df.table.cols.index.is_indexed is True
assert store._handle.root.df.table.cols.B.is_indexed is True
# data column searching
result = store.select("df", "B>0")
expected = df[df.B > 0]
tm.assert_frame_equal(result, expected)
# data column searching (with an indexable and a data_columns)
result = store.select("df", "B>0 and index>df.index[3]")
df_new = df.reindex(index=df.index[4:])
expected = df_new[df_new.B > 0]
tm.assert_frame_equal(result, expected)
# data column selection with a string data_column
df_new = df.copy()
df_new["string"] = "foo"
df_new.loc[1:4, "string"] = np.nan
df_new.loc[5:6, "string"] = "bar"
_maybe_remove(store, "df")
store.append("df", df_new, data_columns=["string"])
result = store.select("df", "string='foo'")
expected = df_new[df_new.string == "foo"]
tm.assert_frame_equal(result, expected)
# using min_itemsize and a data column
def check_col(key, name, size):
assert (
getattr(store.get_storer(key).table.description, name).itemsize
== size
)
with ensure_clean_store(setup_path) as store:
_maybe_remove(store, "df")
store.append(
"df", df_new, data_columns=["string"], min_itemsize={"string": 30}
)
check_col("df", "string", 30)
_maybe_remove(store, "df")
store.append("df", df_new, data_columns=["string"], min_itemsize=30)
check_col("df", "string", 30)
_maybe_remove(store, "df")
store.append(
"df", df_new, data_columns=["string"], min_itemsize={"values": 30}
)
check_col("df", "string", 30)
with ensure_clean_store(setup_path) as store:
df_new["string2"] = "foobarbah"
df_new["string_block1"] = "foobarbah1"
df_new["string_block2"] = "foobarbah2"
_maybe_remove(store, "df")
store.append(
"df",
df_new,
data_columns=["string", "string2"],
min_itemsize={"string": 30, "string2": 40, "values": 50},
)
check_col("df", "string", 30)
check_col("df", "string2", 40)
check_col("df", "values_block_1", 50)
with ensure_clean_store(setup_path) as store:
# multiple data columns
df_new = df.copy()
df_new.iloc[0, df_new.columns.get_loc("A")] = 1.0
df_new.iloc[0, df_new.columns.get_loc("B")] = -1.0
df_new["string"] = "foo"
sl = df_new.columns.get_loc("string")
df_new.iloc[1:4, sl] = np.nan
df_new.iloc[5:6, sl] = "bar"
df_new["string2"] = "foo"
sl = df_new.columns.get_loc("string2")
df_new.iloc[2:5, sl] = np.nan
df_new.iloc[7:8, sl] = "bar"
_maybe_remove(store, "df")
store.append("df", df_new, data_columns=["A", "B", "string", "string2"])
result = store.select(
"df", "string='foo' and string2='foo' and A>0 and B<0"
)
expected = df_new[
(df_new.string == "foo")
& (df_new.string2 == "foo")
& (df_new.A > 0)
& (df_new.B < 0)
]
tm.assert_frame_equal(result, expected, check_index_type=False)
# yield an empty frame
result = store.select("df", "string='foo' and string2='cool'")
expected = df_new[(df_new.string == "foo") & (df_new.string2 == "cool")]
tm.assert_frame_equal(result, expected, check_index_type=False)
with ensure_clean_store(setup_path) as store:
# doc example
df_dc = df.copy()
df_dc["string"] = "foo"
df_dc.loc[4:6, "string"] = np.nan
df_dc.loc[7:9, "string"] = "bar"
df_dc["string2"] = "cool"
df_dc["datetime"] = Timestamp("20010102")
df_dc = df_dc._convert(datetime=True)
df_dc.loc[3:5, ["A", "B", "datetime"]] = np.nan
_maybe_remove(store, "df_dc")
store.append(
"df_dc", df_dc, data_columns=["B", "C", "string", "string2", "datetime"]
)
result = store.select("df_dc", "B>0")
expected = df_dc[df_dc.B > 0]
tm.assert_frame_equal(result, expected, check_index_type=False)
result = store.select("df_dc", ["B > 0", "C > 0", "string == foo"])
expected = df_dc[(df_dc.B > 0) & (df_dc.C > 0) & (df_dc.string == "foo")]
tm.assert_frame_equal(result, expected, check_index_type=False)
with ensure_clean_store(setup_path) as store:
# doc example part 2
np.random.seed(1234)
index = date_range("1/1/2000", periods=8)
df_dc = DataFrame(
np.random.randn(8, 3), index=index, columns=["A", "B", "C"]
)
df_dc["string"] = "foo"
df_dc.loc[4:6, "string"] = np.nan
df_dc.loc[7:9, "string"] = "bar"
df_dc.loc[:, ["B", "C"]] = df_dc.loc[:, ["B", "C"]].abs()
df_dc["string2"] = "cool"
# on-disk operations
store.append("df_dc", df_dc, data_columns=["B", "C", "string", "string2"])
result = store.select("df_dc", "B>0")
expected = df_dc[df_dc.B > 0]
tm.assert_frame_equal(result, expected)
result = store.select("df_dc", ["B > 0", "C > 0", 'string == "foo"'])
expected = df_dc[(df_dc.B > 0) & (df_dc.C > 0) & (df_dc.string == "foo")]
tm.assert_frame_equal(result, expected)
def test_create_table_index(self, setup_path):
with ensure_clean_store(setup_path) as store:
with catch_warnings(record=True):
def col(t, column):
return getattr(store.get_storer(t).table.cols, column)
# data columns
df = tm.makeTimeDataFrame()
df["string"] = "foo"
df["string2"] = "bar"
store.append("f", df, data_columns=["string", "string2"])
assert col("f", "index").is_indexed is True
assert col("f", "string").is_indexed is True
assert col("f", "string2").is_indexed is True
# specify index=columns
store.append(
"f2", df, index=["string"], data_columns=["string", "string2"]
)
assert col("f2", "index").is_indexed is False
assert col("f2", "string").is_indexed is True
assert col("f2", "string2").is_indexed is False
# try to index a non-table
_maybe_remove(store, "f2")
store.put("f2", df)
with pytest.raises(TypeError):
store.create_table_index("f2")
def test_append_hierarchical(self, setup_path):
index = MultiIndex(
levels=[["foo", "bar", "baz", "qux"], ["one", "two", "three"]],
codes=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3], [0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
names=["foo", "bar"],
)
df = DataFrame(np.random.randn(10, 3), index=index, columns=["A", "B", "C"])
with ensure_clean_store(setup_path) as store:
store.append("mi", df)
result = store.select("mi")
tm.assert_frame_equal(result, df)
# GH 3748
result = store.select("mi", columns=["A", "B"])
expected = df.reindex(columns=["A", "B"])
tm.assert_frame_equal(result, expected)
with ensure_clean_path("test.hdf") as path:
df.to_hdf(path, "df", format="table")
result = read_hdf(path, "df", columns=["A", "B"])
expected = df.reindex(columns=["A", "B"])
tm.assert_frame_equal(result, expected)
def test_column_multiindex(self, setup_path):
# GH 4710
# recreate multi-indexes properly
index = MultiIndex.from_tuples(
[("A", "a"), ("A", "b"), ("B", "a"), ("B", "b")], names=["first", "second"]
)
df = DataFrame(np.arange(12).reshape(3, 4), columns=index)
expected = df.copy()
if isinstance(expected.index, RangeIndex):
expected.index = Int64Index(expected.index)
with ensure_clean_store(setup_path) as store:
store.put("df", df)
tm.assert_frame_equal(
store["df"], expected, check_index_type=True, check_column_type=True
)
store.put("df1", df, format="table")
tm.assert_frame_equal(
store["df1"], expected, check_index_type=True, check_column_type=True
)
with pytest.raises(ValueError):
store.put("df2", df, format="table", data_columns=["A"])
with pytest.raises(ValueError):
store.put("df3", df, format="table", data_columns=True)
# appending multi-column on existing table (see GH 6167)
with ensure_clean_store(setup_path) as store:
store.append("df2", df)
store.append("df2", df)
tm.assert_frame_equal(store["df2"], concat((df, df)))
# non_index_axes name
df = DataFrame(
np.arange(12).reshape(3, 4), columns=Index(list("ABCD"), name="foo")
)
expected = df.copy()
if isinstance(expected.index, RangeIndex):
expected.index = Int64Index(expected.index)
with ensure_clean_store(setup_path) as store:
store.put("df1", df, format="table")
tm.assert_frame_equal(
store["df1"], expected, check_index_type=True, check_column_type=True
)
def test_store_multiindex(self, setup_path):
# validate multi-index names
# GH 5527
with ensure_clean_store(setup_path) as store:
def make_index(names=None):
return MultiIndex.from_tuples(
[
(datetime.datetime(2013, 12, d), s, t)
for d in range(1, 3)
for s in range(2)
for t in range(3)
],
names=names,
)
# no names
_maybe_remove(store, "df")
df = DataFrame(np.zeros((12, 2)), columns=["a", "b"], index=make_index())
store.append("df", df)
tm.assert_frame_equal(store.select("df"), df)
# partial names
_maybe_remove(store, "df")
df = DataFrame(
np.zeros((12, 2)),
columns=["a", "b"],
index=make_index(["date", None, None]),
)
store.append("df", df)
tm.assert_frame_equal(store.select("df"), df)
# series
_maybe_remove(store, "s")
s = Series(np.zeros(12), index=make_index(["date", None, None]))
store.append("s", s)
xp = Series(np.zeros(12), index=make_index(["date", "level_1", "level_2"]))
tm.assert_series_equal(store.select("s"), xp)
# dup with column
_maybe_remove(store, "df")
df = DataFrame(
np.zeros((12, 2)),
columns=["a", "b"],
index=make_index(["date", "a", "t"]),
)
with pytest.raises(ValueError):
store.append("df", df)
# dup within level
_maybe_remove(store, "df")
df = DataFrame(
np.zeros((12, 2)),
columns=["a", "b"],
index=make_index(["date", "date", "date"]),
)
with pytest.raises(ValueError):
store.append("df", df)
# fully names
_maybe_remove(store, "df")
df = DataFrame(
np.zeros((12, 2)),
columns=["a", "b"],
index=make_index(["date", "s", "t"]),
)
store.append("df", df)
tm.assert_frame_equal(store.select("df"), df)
def test_select_columns_in_where(self, setup_path):
# GH 6169
# recreate multi-indexes when columns is passed
# in the `where` argument
index = MultiIndex(
levels=[["foo", "bar", "baz", "qux"], ["one", "two", "three"]],
codes=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3], [0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
names=["foo_name", "bar_name"],
)
# With a DataFrame
df = DataFrame(np.random.randn(10, 3), index=index, columns=["A", "B", "C"])
with ensure_clean_store(setup_path) as store:
store.put("df", df, format="table")
expected = df[["A"]]
tm.assert_frame_equal(store.select("df", columns=["A"]), expected)
tm.assert_frame_equal(store.select("df", where="columns=['A']"), expected)
# With a Series
s = Series(np.random.randn(10), index=index, name="A")
with ensure_clean_store(setup_path) as store:
store.put("s", s, format="table")
tm.assert_series_equal(store.select("s", where="columns=['A']"), s)
def test_mi_data_columns(self, setup_path):
# GH 14435
idx = pd.MultiIndex.from_arrays(
[date_range("2000-01-01", periods=5), range(5)], names=["date", "id"]
)
df = pd.DataFrame({"a": [1.1, 1.2, 1.3, 1.4, 1.5]}, index=idx)
with ensure_clean_store(setup_path) as store:
store.append("df", df, data_columns=True)
actual = store.select("df", where="id == 1")
expected = df.iloc[[1], :]
tm.assert_frame_equal(actual, expected)
def test_pass_spec_to_storer(self, setup_path):
df = tm.makeDataFrame()
with ensure_clean_store(setup_path) as store:
store.put("df", df)
with pytest.raises(TypeError):
store.select("df", columns=["A"])
with pytest.raises(TypeError):
store.select("df", where=[("columns=A")])
@td.xfail_non_writeable
def test_append_misc(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = tm.makeDataFrame()
store.append("df", df, chunksize=1)
result = store.select("df")
tm.assert_frame_equal(result, df)
store.append("df1", df, expectedrows=10)
result = store.select("df1")
tm.assert_frame_equal(result, df)
# more chunksize in append tests
def check(obj, comparator):
for c in [10, 200, 1000]:
with ensure_clean_store(setup_path, mode="w") as store:
store.append("obj", obj, chunksize=c)
result = store.select("obj")
comparator(result, obj)
df = tm.makeDataFrame()
df["string"] = "foo"
df["float322"] = 1.0
df["float322"] = df["float322"].astype("float32")
df["bool"] = df["float322"] > 0
df["time1"] = Timestamp("20130101")
df["time2"] = Timestamp("20130102")
check(df, tm.assert_frame_equal)
# empty frame, GH4273
with ensure_clean_store(setup_path) as store:
# 0 len
df_empty = DataFrame(columns=list("ABC"))
store.append("df", df_empty)
with pytest.raises(KeyError, match="'No object named df in the file'"):
store.select("df")
# repeated append of 0/non-zero frames
df = DataFrame(np.random.rand(10, 3), columns=list("ABC"))
store.append("df", df)
tm.assert_frame_equal(store.select("df"), df)
store.append("df", df_empty)
tm.assert_frame_equal(store.select("df"), df)
# store
df = DataFrame(columns=list("ABC"))
store.put("df2", df)
tm.assert_frame_equal(store.select("df2"), df)
def test_append_raise(self, setup_path):
with ensure_clean_store(setup_path) as store:
# test append with invalid input to get good error messages
# list in column
df = tm.makeDataFrame()
df["invalid"] = [["a"]] * len(df)
assert df.dtypes["invalid"] == np.object_
with pytest.raises(TypeError):
store.append("df", df)
# multiple invalid columns
df["invalid2"] = [["a"]] * len(df)
df["invalid3"] = [["a"]] * len(df)
with pytest.raises(TypeError):
store.append("df", df)
# datetime with embedded nans as object
df = tm.makeDataFrame()
s = Series(datetime.datetime(2001, 1, 2), index=df.index)
s = s.astype(object)
s[0:5] = np.nan
df["invalid"] = s
assert df.dtypes["invalid"] == np.object_
with pytest.raises(TypeError):
store.append("df", df)
# directly ndarray
with pytest.raises(TypeError):
store.append("df", np.arange(10))
# series directly
with pytest.raises(TypeError):
store.append("df", Series(np.arange(10)))
# appending an incompatible table
df = tm.makeDataFrame()
store.append("df", df)
df["foo"] = "foo"
with pytest.raises(ValueError):
store.append("df", df)
def test_table_index_incompatible_dtypes(self, setup_path):
df1 = DataFrame({"a": [1, 2, 3]})
df2 = DataFrame({"a": [4, 5, 6]}, index=date_range("1/1/2000", periods=3))
with ensure_clean_store(setup_path) as store:
store.put("frame", df1, format="table")
with pytest.raises(TypeError):
store.put("frame", df2, format="table", append=True)
def test_table_values_dtypes_roundtrip(self, setup_path):
with ensure_clean_store(setup_path) as store:
df1 = DataFrame({"a": [1, 2, 3]}, dtype="f8")
store.append("df_f8", df1)
tm.assert_series_equal(df1.dtypes, store["df_f8"].dtypes)
df2 = DataFrame({"a": [1, 2, 3]}, dtype="i8")
store.append("df_i8", df2)
tm.assert_series_equal(df2.dtypes, store["df_i8"].dtypes)
# incompatible dtype
with pytest.raises(ValueError):
store.append("df_i8", df1)
# check creation/storage/retrieval of float32 (a bit hacky to
# actually create them thought)
df1 = DataFrame(np.array([[1], [2], [3]], dtype="f4"), columns=["A"])
store.append("df_f4", df1)
tm.assert_series_equal(df1.dtypes, store["df_f4"].dtypes)
assert df1.dtypes[0] == "float32"
# check with mixed dtypes
df1 = DataFrame(
{
c: Series(np.random.randint(5), dtype=c)
for c in ["float32", "float64", "int32", "int64", "int16", "int8"]
}
)
df1["string"] = "foo"
df1["float322"] = 1.0
df1["float322"] = df1["float322"].astype("float32")
df1["bool"] = df1["float32"] > 0
df1["time1"] = Timestamp("20130101")
df1["time2"] = Timestamp("20130102")
store.append("df_mixed_dtypes1", df1)
result = store.select("df_mixed_dtypes1").dtypes.value_counts()
result.index = [str(i) for i in result.index]
expected = Series(
{
"float32": 2,
"float64": 1,
"int32": 1,
"bool": 1,
"int16": 1,
"int8": 1,
"int64": 1,
"object": 1,
"datetime64[ns]": 2,
}
)
result = result.sort_index()
expected = expected.sort_index()
tm.assert_series_equal(result, expected)
def test_table_mixed_dtypes(self, setup_path):
# frame
df = tm.makeDataFrame()
df["obj1"] = "foo"
df["obj2"] = "bar"
df["bool1"] = df["A"] > 0
df["bool2"] = df["B"] > 0
df["bool3"] = True
df["int1"] = 1
df["int2"] = 2
df["timestamp1"] = Timestamp("20010102")
df["timestamp2"] = Timestamp("20010103")
df["datetime1"] = datetime.datetime(2001, 1, 2, 0, 0)
df["datetime2"] = datetime.datetime(2001, 1, 3, 0, 0)
df.loc[3:6, ["obj1"]] = np.nan
df = df._consolidate()._convert(datetime=True)
with ensure_clean_store(setup_path) as store:
store.append("df1_mixed", df)
tm.assert_frame_equal(store.select("df1_mixed"), df)
def test_unimplemented_dtypes_table_columns(self, setup_path):
with ensure_clean_store(setup_path) as store:
dtypes = [("date", datetime.date(2001, 1, 2))]
# currently not supported dtypes ####
for n, f in dtypes:
df = tm.makeDataFrame()
df[n] = f
with pytest.raises(TypeError):
store.append("df1_{n}".format(n=n), df)
# frame
df = tm.makeDataFrame()
df["obj1"] = "foo"
df["obj2"] = "bar"
df["datetime1"] = datetime.date(2001, 1, 2)
df = df._consolidate()._convert(datetime=True)
with ensure_clean_store(setup_path) as store:
# this fails because we have a date in the object block......
with pytest.raises(TypeError):
store.append("df_unimplemented", df)
@td.xfail_non_writeable
@pytest.mark.skipif(
LooseVersion(np.__version__) == LooseVersion("1.15.0"),
reason=(
"Skipping pytables test when numpy version is "
"exactly equal to 1.15.0: gh-22098"
),
)
def test_calendar_roundtrip_issue(self, setup_path):
# 8591
# doc example from tseries holiday section
weekmask_egypt = "Sun Mon Tue Wed Thu"
holidays = [
"2012-05-01",
datetime.datetime(2013, 5, 1),
np.datetime64("2014-05-01"),
]
bday_egypt = pd.offsets.CustomBusinessDay(
holidays=holidays, weekmask=weekmask_egypt
)
dt = datetime.datetime(2013, 4, 30)
dts = date_range(dt, periods=5, freq=bday_egypt)
s = Series(dts.weekday, dts).map(Series("Mon Tue Wed Thu Fri Sat Sun".split()))
with ensure_clean_store(setup_path) as store:
store.put("fixed", s)
result = store.select("fixed")
tm.assert_series_equal(result, s)
store.append("table", s)
result = store.select("table")
tm.assert_series_equal(result, s)
def test_roundtrip_tz_aware_index(self, setup_path):
# GH 17618
time = pd.Timestamp("2000-01-01 01:00:00", tz="US/Eastern")
df = pd.DataFrame(data=[0], index=[time])
with ensure_clean_store(setup_path) as store:
store.put("frame", df, format="fixed")
recons = store["frame"]
tm.assert_frame_equal(recons, df)
assert recons.index[0].value == 946706400000000000
def test_append_with_timedelta(self, setup_path):
# GH 3577
# append timedelta
df = DataFrame(
dict(
A=Timestamp("20130101"),
B=[
Timestamp("20130101") + timedelta(days=i, seconds=10)
for i in range(10)
],
)
)
df["C"] = df["A"] - df["B"]
df.loc[3:5, "C"] = np.nan
with ensure_clean_store(setup_path) as store:
# table
_maybe_remove(store, "df")
store.append("df", df, data_columns=True)
result = store.select("df")
tm.assert_frame_equal(result, df)
result = store.select("df", where="C<100000")
tm.assert_frame_equal(result, df)
result = store.select("df", where="C<pd.Timedelta('-3D')")
tm.assert_frame_equal(result, df.iloc[3:])
result = store.select("df", "C<'-3D'")
tm.assert_frame_equal(result, df.iloc[3:])
# a bit hacky here as we don't really deal with the NaT properly
result = store.select("df", "C<'-500000s'")
result = result.dropna(subset=["C"])
tm.assert_frame_equal(result, df.iloc[6:])
result = store.select("df", "C<'-3.5D'")
result = result.iloc[1:]
tm.assert_frame_equal(result, df.iloc[4:])
# fixed
_maybe_remove(store, "df2")
store.put("df2", df)
result = store.select("df2")
tm.assert_frame_equal(result, df)
def test_remove(self, setup_path):
with ensure_clean_store(setup_path) as store:
ts = tm.makeTimeSeries()
df = tm.makeDataFrame()
store["a"] = ts
store["b"] = df
_maybe_remove(store, "a")
assert len(store) == 1
tm.assert_frame_equal(df, store["b"])
_maybe_remove(store, "b")
assert len(store) == 0
# nonexistence
with pytest.raises(
KeyError, match="'No object named a_nonexistent_store in the file'"
):
store.remove("a_nonexistent_store")
# pathing
store["a"] = ts
store["b/foo"] = df
_maybe_remove(store, "foo")
_maybe_remove(store, "b/foo")
assert len(store) == 1
store["a"] = ts
store["b/foo"] = df
_maybe_remove(store, "b")
assert len(store) == 1
# __delitem__
store["a"] = ts
store["b"] = df
del store["a"]
del store["b"]
assert len(store) == 0
def test_invalid_terms(self, setup_path):
with ensure_clean_store(setup_path) as store:
with catch_warnings(record=True):
df = tm.makeTimeDataFrame()
df["string"] = "foo"
df.loc[0:4, "string"] = "bar"
store.put("df", df, format="table")
# some invalid terms
with pytest.raises(TypeError):
Term()
# more invalid
with pytest.raises(ValueError):
store.select("df", "df.index[3]")
with pytest.raises(SyntaxError):
store.select("df", "index>")
# from the docs
with ensure_clean_path(setup_path) as path:
dfq = DataFrame(
np.random.randn(10, 4),
columns=list("ABCD"),
index=date_range("20130101", periods=10),
)
dfq.to_hdf(path, "dfq", format="table", data_columns=True)
# check ok
read_hdf(
path, "dfq", where="index>Timestamp('20130104') & columns=['A', 'B']"
)
read_hdf(path, "dfq", where="A>0 or C>0")
# catch the invalid reference
with ensure_clean_path(setup_path) as path:
dfq = DataFrame(
np.random.randn(10, 4),
columns=list("ABCD"),
index=date_range("20130101", periods=10),
)
dfq.to_hdf(path, "dfq", format="table")
with pytest.raises(ValueError):
read_hdf(path, "dfq", where="A>0 or C>0")
def test_same_name_scoping(self, setup_path):
with ensure_clean_store(setup_path) as store:
import pandas as pd
df = DataFrame(
np.random.randn(20, 2), index=pd.date_range("20130101", periods=20)
)
store.put("df", df, format="table")
expected = df[df.index > pd.Timestamp("20130105")]
import datetime # noqa
result = store.select("df", "index>datetime.datetime(2013,1,5)")
tm.assert_frame_equal(result, expected)
from datetime import datetime # noqa
# technically an error, but allow it
result = store.select("df", "index>datetime.datetime(2013,1,5)")
tm.assert_frame_equal(result, expected)
result = store.select("df", "index>datetime(2013,1,5)")
tm.assert_frame_equal(result, expected)
def test_series(self, setup_path):
s = tm.makeStringSeries()
self._check_roundtrip(s, tm.assert_series_equal, path=setup_path)
ts = tm.makeTimeSeries()
self._check_roundtrip(ts, tm.assert_series_equal, path=setup_path)
ts2 = Series(ts.index, Index(ts.index, dtype=object))
self._check_roundtrip(ts2, tm.assert_series_equal, path=setup_path)
ts3 = Series(ts.values, Index(np.asarray(ts.index, dtype=object), dtype=object))
self._check_roundtrip(
ts3, tm.assert_series_equal, path=setup_path, check_index_type=False
)
def test_float_index(self, setup_path):
# GH #454
index = np.random.randn(10)
s = Series(np.random.randn(10), index=index)
self._check_roundtrip(s, tm.assert_series_equal, path=setup_path)
@td.xfail_non_writeable
def test_tuple_index(self, setup_path):
# GH #492
col = np.arange(10)
idx = [(0.0, 1.0), (2.0, 3.0), (4.0, 5.0)]
data = np.random.randn(30).reshape((3, 10))
DF = DataFrame(data, index=idx, columns=col)
with catch_warnings(record=True):
simplefilter("ignore", pd.errors.PerformanceWarning)
self._check_roundtrip(DF, tm.assert_frame_equal, path=setup_path)
@td.xfail_non_writeable
@pytest.mark.filterwarnings("ignore::pandas.errors.PerformanceWarning")
def test_index_types(self, setup_path):
with catch_warnings(record=True):
values = np.random.randn(2)
func = lambda l, r: tm.assert_series_equal(
l, r, check_dtype=True, check_index_type=True, check_series_type=True
)
with catch_warnings(record=True):
ser = Series(values, [0, "y"])
self._check_roundtrip(ser, func, path=setup_path)
with catch_warnings(record=True):
ser = Series(values, [datetime.datetime.today(), 0])
self._check_roundtrip(ser, func, path=setup_path)
with catch_warnings(record=True):
ser = Series(values, ["y", 0])
self._check_roundtrip(ser, func, path=setup_path)
with catch_warnings(record=True):
ser = Series(values, [datetime.date.today(), "a"])
self._check_roundtrip(ser, func, path=setup_path)
with catch_warnings(record=True):
ser = Series(values, [0, "y"])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(values, [datetime.datetime.today(), 0])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(values, ["y", 0])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(values, [datetime.date.today(), "a"])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(values, [1.23, "b"])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(values, [1, 1.53])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(values, [1, 5])
self._check_roundtrip(ser, func, path=setup_path)
ser = Series(
values, [datetime.datetime(2012, 1, 1), datetime.datetime(2012, 1, 2)]
)
self._check_roundtrip(ser, func, path=setup_path)
def test_timeseries_preepoch(self, setup_path):
dr = bdate_range("1/1/1940", "1/1/1960")
ts = Series(np.random.randn(len(dr)), index=dr)
try:
self._check_roundtrip(ts, tm.assert_series_equal, path=setup_path)
except OverflowError:
pytest.skip("known failer on some windows platforms")
@td.xfail_non_writeable
@pytest.mark.parametrize(
"compression", [False, pytest.param(True, marks=td.skip_if_windows_python_3)]
)
def test_frame(self, compression, setup_path):
df = tm.makeDataFrame()
# put in some random NAs
df.values[0, 0] = np.nan
df.values[5, 3] = np.nan
self._check_roundtrip_table(
df, tm.assert_frame_equal, path=setup_path, compression=compression
)
self._check_roundtrip(
df, tm.assert_frame_equal, path=setup_path, compression=compression
)
tdf = tm.makeTimeDataFrame()
self._check_roundtrip(
tdf, tm.assert_frame_equal, path=setup_path, compression=compression
)
with ensure_clean_store(setup_path) as store:
# not consolidated
df["foo"] = np.random.randn(len(df))
store["df"] = df
recons = store["df"]
assert recons._data.is_consolidated()
# empty
self._check_roundtrip(df[:0], tm.assert_frame_equal, path=setup_path)
@td.xfail_non_writeable
def test_empty_series_frame(self, setup_path):
s0 = Series(dtype=object)
s1 = Series(name="myseries", dtype=object)
df0 = DataFrame()
df1 = DataFrame(index=["a", "b", "c"])
df2 = DataFrame(columns=["d", "e", "f"])
self._check_roundtrip(s0, tm.assert_series_equal, path=setup_path)
self._check_roundtrip(s1, tm.assert_series_equal, path=setup_path)
self._check_roundtrip(df0, tm.assert_frame_equal, path=setup_path)
self._check_roundtrip(df1, tm.assert_frame_equal, path=setup_path)
self._check_roundtrip(df2, tm.assert_frame_equal, path=setup_path)
@td.xfail_non_writeable
@pytest.mark.parametrize(
"dtype", [np.int64, np.float64, np.object, "m8[ns]", "M8[ns]"]
)
def test_empty_series(self, dtype, setup_path):
s = Series(dtype=dtype)
self._check_roundtrip(s, tm.assert_series_equal, path=setup_path)
def test_can_serialize_dates(self, setup_path):
rng = [x.date() for x in bdate_range("1/1/2000", "1/30/2000")]
frame = DataFrame(np.random.randn(len(rng), 4), index=rng)
self._check_roundtrip(frame, tm.assert_frame_equal, path=setup_path)
def test_store_hierarchical(self, setup_path):
index = MultiIndex(
levels=[["foo", "bar", "baz", "qux"], ["one", "two", "three"]],
codes=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3], [0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
names=["foo", "bar"],
)
frame = DataFrame(np.random.randn(10, 3), index=index, columns=["A", "B", "C"])
self._check_roundtrip(frame, tm.assert_frame_equal, path=setup_path)
self._check_roundtrip(frame.T, tm.assert_frame_equal, path=setup_path)
self._check_roundtrip(frame["A"], tm.assert_series_equal, path=setup_path)
# check that the names are stored
with ensure_clean_store(setup_path) as store:
store["frame"] = frame
recons = store["frame"]
tm.assert_frame_equal(recons, frame)
def test_store_index_name(self, setup_path):
df = tm.makeDataFrame()
df.index.name = "foo"
with ensure_clean_store(setup_path) as store:
store["frame"] = df
recons = store["frame"]
tm.assert_frame_equal(recons, df)
def test_store_index_name_with_tz(self, setup_path):
# GH 13884
df = pd.DataFrame({"A": [1, 2]})
df.index = pd.DatetimeIndex([1234567890123456787, 1234567890123456788])
df.index = df.index.tz_localize("UTC")
df.index.name = "foo"
with ensure_clean_store(setup_path) as store:
store.put("frame", df, format="table")
recons = store["frame"]
tm.assert_frame_equal(recons, df)
@pytest.mark.parametrize("table_format", ["table", "fixed"])
def test_store_index_name_numpy_str(self, table_format, setup_path):
# GH #13492
idx = pd.Index(
pd.to_datetime([datetime.date(2000, 1, 1), datetime.date(2000, 1, 2)]),
name="cols\u05d2",
)
idx1 = pd.Index(
pd.to_datetime([datetime.date(2010, 1, 1), datetime.date(2010, 1, 2)]),
name="rows\u05d0",
)
df = pd.DataFrame(np.arange(4).reshape(2, 2), columns=idx, index=idx1)
# This used to fail, returning numpy strings instead of python strings.
with ensure_clean_path(setup_path) as path:
df.to_hdf(path, "df", format=table_format)
df2 = read_hdf(path, "df")
tm.assert_frame_equal(df, df2, check_names=True)
assert type(df2.index.name) == str
assert type(df2.columns.name) == str
def test_store_series_name(self, setup_path):
df = tm.makeDataFrame()
series = df["A"]
with ensure_clean_store(setup_path) as store:
store["series"] = series
recons = store["series"]
tm.assert_series_equal(recons, series)
@td.xfail_non_writeable
@pytest.mark.parametrize(
"compression", [False, pytest.param(True, marks=td.skip_if_windows_python_3)]
)
def test_store_mixed(self, compression, setup_path):
def _make_one():
df = tm.makeDataFrame()
df["obj1"] = "foo"
df["obj2"] = "bar"
df["bool1"] = df["A"] > 0
df["bool2"] = df["B"] > 0
df["int1"] = 1
df["int2"] = 2
return df._consolidate()
df1 = _make_one()
df2 = _make_one()
self._check_roundtrip(df1, tm.assert_frame_equal, path=setup_path)
self._check_roundtrip(df2, tm.assert_frame_equal, path=setup_path)
with ensure_clean_store(setup_path) as store:
store["obj"] = df1
tm.assert_frame_equal(store["obj"], df1)
store["obj"] = df2
tm.assert_frame_equal(store["obj"], df2)
# check that can store Series of all of these types
self._check_roundtrip(
df1["obj1"],
tm.assert_series_equal,
path=setup_path,
compression=compression,
)
self._check_roundtrip(
df1["bool1"],
tm.assert_series_equal,
path=setup_path,
compression=compression,
)
self._check_roundtrip(
df1["int1"],
tm.assert_series_equal,
path=setup_path,
compression=compression,
)
@pytest.mark.filterwarnings(
"ignore:\\nduplicate:pandas.io.pytables.DuplicateWarning"
)
def test_select_with_dups(self, setup_path):
# single dtypes
df = DataFrame(np.random.randn(10, 4), columns=["A", "A", "B", "B"])
df.index = date_range("20130101 9:30", periods=10, freq="T")
with ensure_clean_store(setup_path) as store:
store.append("df", df)
result = store.select("df")
expected = df
tm.assert_frame_equal(result, expected, by_blocks=True)
result = store.select("df", columns=df.columns)
expected = df
tm.assert_frame_equal(result, expected, by_blocks=True)
result = store.select("df", columns=["A"])
expected = df.loc[:, ["A"]]
tm.assert_frame_equal(result, expected)
# dups across dtypes
df = concat(
[
DataFrame(np.random.randn(10, 4), columns=["A", "A", "B", "B"]),
DataFrame(
np.random.randint(0, 10, size=20).reshape(10, 2), columns=["A", "C"]
),
],
axis=1,
)
df.index = date_range("20130101 9:30", periods=10, freq="T")
with ensure_clean_store(setup_path) as store:
store.append("df", df)
result = store.select("df")
expected = df
tm.assert_frame_equal(result, expected, by_blocks=True)
result = store.select("df", columns=df.columns)
expected = df
tm.assert_frame_equal(result, expected, by_blocks=True)
expected = df.loc[:, ["A"]]
result = store.select("df", columns=["A"])
tm.assert_frame_equal(result, expected, by_blocks=True)
expected = df.loc[:, ["B", "A"]]
result = store.select("df", columns=["B", "A"])
tm.assert_frame_equal(result, expected, by_blocks=True)
# duplicates on both index and columns
with ensure_clean_store(setup_path) as store:
store.append("df", df)
store.append("df", df)
expected = df.loc[:, ["B", "A"]]
expected = concat([expected, expected])
result = store.select("df", columns=["B", "A"])
tm.assert_frame_equal(result, expected, by_blocks=True)
def test_overwrite_node(self, setup_path):
with ensure_clean_store(setup_path) as store:
store["a"] = tm.makeTimeDataFrame()
ts = tm.makeTimeSeries()
store["a"] = ts
tm.assert_series_equal(store["a"], ts)
def test_select(self, setup_path):
with ensure_clean_store(setup_path) as store:
with catch_warnings(record=True):
# select with columns=
df = tm.makeTimeDataFrame()
_maybe_remove(store, "df")
store.append("df", df)
result = store.select("df", columns=["A", "B"])
expected = df.reindex(columns=["A", "B"])
tm.assert_frame_equal(expected, result)
# equivalently
result = store.select("df", [("columns=['A', 'B']")])
expected = df.reindex(columns=["A", "B"])
tm.assert_frame_equal(expected, result)
# with a data column
_maybe_remove(store, "df")
store.append("df", df, data_columns=["A"])
result = store.select("df", ["A > 0"], columns=["A", "B"])
expected = df[df.A > 0].reindex(columns=["A", "B"])
tm.assert_frame_equal(expected, result)
# all a data columns
_maybe_remove(store, "df")
store.append("df", df, data_columns=True)
result = store.select("df", ["A > 0"], columns=["A", "B"])
expected = df[df.A > 0].reindex(columns=["A", "B"])
tm.assert_frame_equal(expected, result)
# with a data column, but different columns
_maybe_remove(store, "df")
store.append("df", df, data_columns=["A"])
result = store.select("df", ["A > 0"], columns=["C", "D"])
expected = df[df.A > 0].reindex(columns=["C", "D"])
tm.assert_frame_equal(expected, result)
def test_select_dtypes(self, setup_path):
with ensure_clean_store(setup_path) as store:
# with a Timestamp data column (GH #2637)
df = DataFrame(
dict(ts=bdate_range("2012-01-01", periods=300), A=np.random.randn(300))
)
_maybe_remove(store, "df")
store.append("df", df, data_columns=["ts", "A"])
result = store.select("df", "ts>=Timestamp('2012-02-01')")
expected = df[df.ts >= Timestamp("2012-02-01")]
tm.assert_frame_equal(expected, result)
# bool columns (GH #2849)
df = DataFrame(np.random.randn(5, 2), columns=["A", "B"])
df["object"] = "foo"
df.loc[4:5, "object"] = "bar"
df["boolv"] = df["A"] > 0
_maybe_remove(store, "df")
store.append("df", df, data_columns=True)
expected = df[df.boolv == True].reindex(columns=["A", "boolv"]) # noqa
for v in [True, "true", 1]:
result = store.select(
"df", "boolv == {v!s}".format(v=v), columns=["A", "boolv"]
)
tm.assert_frame_equal(expected, result)
expected = df[df.boolv == False].reindex(columns=["A", "boolv"]) # noqa
for v in [False, "false", 0]:
result = store.select(
"df", "boolv == {v!s}".format(v=v), columns=["A", "boolv"]
)
tm.assert_frame_equal(expected, result)
# integer index
df = DataFrame(dict(A=np.random.rand(20), B=np.random.rand(20)))
_maybe_remove(store, "df_int")
store.append("df_int", df)
result = store.select("df_int", "index<10 and columns=['A']")
expected = df.reindex(index=list(df.index)[0:10], columns=["A"])
tm.assert_frame_equal(expected, result)
# float index
df = DataFrame(
dict(
A=np.random.rand(20),
B=np.random.rand(20),
index=np.arange(20, dtype="f8"),
)
)
_maybe_remove(store, "df_float")
store.append("df_float", df)
result = store.select("df_float", "index<10.0 and columns=['A']")
expected = df.reindex(index=list(df.index)[0:10], columns=["A"])
tm.assert_frame_equal(expected, result)
with ensure_clean_store(setup_path) as store:
# floats w/o NaN
df = DataFrame(dict(cols=range(11), values=range(11)), dtype="float64")
df["cols"] = (df["cols"] + 10).apply(str)
store.append("df1", df, data_columns=True)
result = store.select("df1", where="values>2.0")
expected = df[df["values"] > 2.0]
tm.assert_frame_equal(expected, result)
# floats with NaN
df.iloc[0] = np.nan
expected = df[df["values"] > 2.0]
store.append("df2", df, data_columns=True, index=False)
result = store.select("df2", where="values>2.0")
tm.assert_frame_equal(expected, result)
# https://github.com/PyTables/PyTables/issues/282
# bug in selection when 0th row has a np.nan and an index
# store.append('df3',df,data_columns=True)
# result = store.select(
# 'df3', where='values>2.0')
# tm.assert_frame_equal(expected, result)
# not in first position float with NaN ok too
df = DataFrame(dict(cols=range(11), values=range(11)), dtype="float64")
df["cols"] = (df["cols"] + 10).apply(str)
df.iloc[1] = np.nan
expected = df[df["values"] > 2.0]
store.append("df4", df, data_columns=True)
result = store.select("df4", where="values>2.0")
tm.assert_frame_equal(expected, result)
# test selection with comparison against numpy scalar
# GH 11283
with ensure_clean_store(setup_path) as store:
df = tm.makeDataFrame()
expected = df[df["A"] > 0]
store.append("df", df, data_columns=True)
np_zero = np.float64(0) # noqa
result = store.select("df", where=["A>np_zero"])
tm.assert_frame_equal(expected, result)
def test_select_with_many_inputs(self, setup_path):
with ensure_clean_store(setup_path) as store:
df = DataFrame(
dict(
ts=bdate_range("2012-01-01", periods=300),
A=np.random.randn(300),
B=range(300),
users=["a"] * 50
+ ["b"] * 50
+ ["c"] * 100
+ ["a{i:03d}".format(i=i) for i in range(100)],
)
)
_maybe_remove(store, "df")
store.append("df", df, data_columns=["ts", "A", "B", "users"])
# regular select
result = store.select("df", "ts>=Timestamp('2012-02-01')")
expected = df[df.ts >= Timestamp("2012-02-01")]
tm.assert_frame_equal(expected, result)
# small selector
result = store.select(
"df", "ts>=Timestamp('2012-02-01') & users=['a','b','c']"
)
expected = df[
(df.ts >= Timestamp("2012-02-01")) & df.users.isin(["a", "b", "c"])
]
tm.assert_frame_equal(expected, result)
# big selector along the columns
selector = ["a", "b", "c"] + ["a{i:03d}".format(i=i) for i in range(60)]
result = store.select(
"df", "ts>=Timestamp('2012-02-01') and users=selector"
)
expected = df[(df.ts >= Timestamp("2012-02-01")) & df.users.isin(selector)]
tm.assert_frame_equal(expected, result)
selector = range(100, 200)
result = store.select("df", "B=selector")
expected = df[df.B.isin(selector)]
tm.assert_frame_equal(expected, result)
assert len(result) == 100
# big selector along the index
selector = Index(df.ts[0:100].values)
result = store.select("df", "ts=selector")
expected = df[df.ts.isin(selector.values)]
tm.assert_frame_equal(expected, result)
assert len(result) == 100
def test_select_iterator(self, setup_path):
# single table
with ensure_clean_store(setup_path) as store:
df = tm.makeTimeDataFrame(500)
_maybe_remove(store, "df")
store.append("df", df)
expected = store.select("df")
results = list(store.select("df", iterator=True))
result = concat(results)
| tm.assert_frame_equal(expected, result) | pandas.util.testing.assert_frame_equal |
import os
from pathlib import Path
import pandas as pd
import requests
class OisManager:
TOIS_CSV_URL = 'https://exofop.ipac.caltech.edu/tess/download_toi.php?sort=toi&output=csv'
CTOIS_CSV_URL = 'https://exofop.ipac.caltech.edu/tess/download_ctoi.php?sort=ctoi&output=csv'
KOIS_LIST_URL = 'https://exofop.ipac.caltech.edu/kepler/targets.php?sort=num-pc&page1=1&ipp1=100000&koi1=&koi2='
KIC_STAR_URL = 'https://exoplanetarchive.ipac.caltech.edu/cgi-bin/nstedAPI/nph-nstedAPI?table=keplerstellar&select=kepid,dist'
KOI_CSV_URL = 'https://exoplanetarchive.ipac.caltech.edu/cgi-bin/nstedAPI/nph-nstedAPI?table=cumulative'
EPIC_CSV_URL = 'https://exoplanetarchive.ipac.caltech.edu/cgi-bin/nstedAPI/nph-nstedAPI?table=k2candidates&' \
'select=epic_name,epic_candname,k2c_disp,pl_orbper,st_dist,st_teff,st_logg,st_metfe,st_metratio,' \
'st_vsini,st_kep,pl_trandep,pl_trandur,pl_rade,pl_eqt,pl_orbincl,ra_str,dec_str,pl_tranmid'
TOIS_CSV = 'tois.csv'
CTOIS_CSV = 'ctois.csv'
KOIS_CSV = 'kois.csv'
EPIC_CSV = 'epic_ois.csv'
KIC_STAR_CSV = 'kic_star.csv'
ois = None
def __init__(self):
home = str(Path.home()) + "/.sherlockpipe/"
if not os.path.exists(home):
os.mkdir(home)
self.tois_csv = home + self.TOIS_CSV
self.ctois_csv = home + self.CTOIS_CSV
self.kois_csv = home + self.KOIS_CSV
self.epic_csv = home + self.EPIC_CSV
self.kic_star_csv = home + self.KIC_STAR_CSV
def load_ois(self):
if not os.path.isfile(self.tois_csv) or not os.path.isfile(self.ctois_csv):
print("TOIs files are not found. Downloading...")
self.update_tic_csvs()
print("TOIs files download is finished!")
toi_data = pd.read_csv(self.tois_csv)
ois = toi_data
ctoi_data = | pd.read_csv(self.ctois_csv) | pandas.read_csv |
"""
Generic data algorithms. This module is experimental at the moment and not
intended for public consumption
"""
from __future__ import annotations
import operator
from textwrap import dedent
from typing import (
TYPE_CHECKING,
Literal,
Union,
cast,
final,
)
from warnings import warn
import numpy as np
from pandas._libs import (
algos,
hashtable as htable,
iNaT,
lib,
)
from pandas._typing import (
AnyArrayLike,
ArrayLike,
DtypeObj,
Scalar,
TakeIndexer,
npt,
)
from pandas.util._decorators import doc
from pandas.core.dtypes.cast import (
construct_1d_object_array_from_listlike,
infer_dtype_from_array,
sanitize_to_nanoseconds,
)
from pandas.core.dtypes.common import (
ensure_float64,
ensure_object,
ensure_platform_int,
is_array_like,
is_bool_dtype,
is_categorical_dtype,
is_complex_dtype,
is_datetime64_dtype,
is_extension_array_dtype,
is_float_dtype,
is_integer,
is_integer_dtype,
is_list_like,
is_numeric_dtype,
is_object_dtype,
is_scalar,
is_timedelta64_dtype,
needs_i8_conversion,
)
from pandas.core.dtypes.dtypes import PandasDtype
from pandas.core.dtypes.generic import (
ABCDatetimeArray,
ABCExtensionArray,
ABCIndex,
ABCMultiIndex,
ABCRangeIndex,
ABCSeries,
ABCTimedeltaArray,
)
from pandas.core.dtypes.missing import (
isna,
na_value_for_dtype,
)
from pandas.core.array_algos.take import take_nd
from pandas.core.construction import (
array as pd_array,
ensure_wrapped_if_datetimelike,
extract_array,
)
from pandas.core.indexers import validate_indices
if TYPE_CHECKING:
from pandas._typing import (
NumpySorter,
NumpyValueArrayLike,
)
from pandas import (
Categorical,
DataFrame,
Index,
Series,
)
from pandas.core.arrays import (
DatetimeArray,
ExtensionArray,
TimedeltaArray,
)
_shared_docs: dict[str, str] = {}
# --------------- #
# dtype access #
# --------------- #
def _ensure_data(values: ArrayLike) -> np.ndarray:
"""
routine to ensure that our data is of the correct
input dtype for lower-level routines
This will coerce:
- ints -> int64
- uint -> uint64
- bool -> uint64 (TODO this should be uint8)
- datetimelike -> i8
- datetime64tz -> i8 (in local tz)
- categorical -> codes
Parameters
----------
values : np.ndarray or ExtensionArray
Returns
-------
np.ndarray
"""
if not isinstance(values, ABCMultiIndex):
# extract_array would raise
values = extract_array(values, extract_numpy=True)
# we check some simple dtypes first
if is_object_dtype(values.dtype):
return ensure_object(np.asarray(values))
elif is_bool_dtype(values.dtype):
if isinstance(values, np.ndarray):
# i.e. actually dtype == np.dtype("bool")
return np.asarray(values).view("uint8")
else:
# i.e. all-bool Categorical, BooleanArray
try:
return np.asarray(values).astype("uint8", copy=False)
except TypeError:
# GH#42107 we have pd.NAs present
return np.asarray(values)
elif is_integer_dtype(values.dtype):
return np.asarray(values)
elif is_float_dtype(values.dtype):
# Note: checking `values.dtype == "float128"` raises on Windows and 32bit
# error: Item "ExtensionDtype" of "Union[Any, ExtensionDtype, dtype[Any]]"
# has no attribute "itemsize"
if values.dtype.itemsize in [2, 12, 16]: # type: ignore[union-attr]
# we dont (yet) have float128 hashtable support
return ensure_float64(values)
return np.asarray(values)
elif is_complex_dtype(values.dtype):
# Incompatible return value type (got "Tuple[Union[Any, ExtensionArray,
# ndarray[Any, Any]], Union[Any, ExtensionDtype]]", expected
# "Tuple[ndarray[Any, Any], Union[dtype[Any], ExtensionDtype]]")
return values # type: ignore[return-value]
# datetimelike
elif needs_i8_conversion(values.dtype):
if isinstance(values, np.ndarray):
values = sanitize_to_nanoseconds(values)
npvalues = values.view("i8")
npvalues = cast(np.ndarray, npvalues)
return npvalues
elif is_categorical_dtype(values.dtype):
values = cast("Categorical", values)
values = values.codes
return values
# we have failed, return object
values = np.asarray(values, dtype=object)
return ensure_object(values)
def _reconstruct_data(
values: ArrayLike, dtype: DtypeObj, original: AnyArrayLike
) -> ArrayLike:
"""
reverse of _ensure_data
Parameters
----------
values : np.ndarray or ExtensionArray
dtype : np.dtype or ExtensionDtype
original : AnyArrayLike
Returns
-------
ExtensionArray or np.ndarray
"""
if isinstance(values, ABCExtensionArray) and values.dtype == dtype:
# Catch DatetimeArray/TimedeltaArray
return values
if not isinstance(dtype, np.dtype):
# i.e. ExtensionDtype
cls = dtype.construct_array_type()
if isinstance(values, cls) and values.dtype == dtype:
return values
values = cls._from_sequence(values)
elif is_bool_dtype(dtype):
values = values.astype(dtype, copy=False)
# we only support object dtypes bool Index
if isinstance(original, ABCIndex):
values = values.astype(object, copy=False)
elif dtype is not None:
if is_datetime64_dtype(dtype):
dtype = np.dtype("datetime64[ns]")
elif is_timedelta64_dtype(dtype):
dtype = np.dtype("timedelta64[ns]")
values = values.astype(dtype, copy=False)
return values
def _ensure_arraylike(values) -> ArrayLike:
"""
ensure that we are arraylike if not already
"""
if not is_array_like(values):
inferred = lib.infer_dtype(values, skipna=False)
if inferred in ["mixed", "string", "mixed-integer"]:
# "mixed-integer" to ensure we do not cast ["ss", 42] to str GH#22160
if isinstance(values, tuple):
values = list(values)
values = construct_1d_object_array_from_listlike(values)
else:
values = np.asarray(values)
return values
_hashtables = {
"complex128": htable.Complex128HashTable,
"complex64": htable.Complex64HashTable,
"float64": htable.Float64HashTable,
"float32": htable.Float32HashTable,
"uint64": htable.UInt64HashTable,
"uint32": htable.UInt32HashTable,
"uint16": htable.UInt16HashTable,
"uint8": htable.UInt8HashTable,
"int64": htable.Int64HashTable,
"int32": htable.Int32HashTable,
"int16": htable.Int16HashTable,
"int8": htable.Int8HashTable,
"string": htable.StringHashTable,
"object": htable.PyObjectHashTable,
}
def _get_hashtable_algo(values: np.ndarray):
"""
Parameters
----------
values : np.ndarray
Returns
-------
htable : HashTable subclass
values : ndarray
"""
values = _ensure_data(values)
ndtype = _check_object_for_strings(values)
htable = _hashtables[ndtype]
return htable, values
def _get_values_for_rank(values: ArrayLike) -> np.ndarray:
if is_categorical_dtype(values):
values = cast("Categorical", values)._values_for_rank()
values = _ensure_data(values)
if values.dtype.kind in ["i", "u", "f"]:
# rank_t includes only object, int64, uint64, float64
dtype = values.dtype.kind + "8"
values = values.astype(dtype, copy=False)
return values
def get_data_algo(values: ArrayLike):
values = _get_values_for_rank(values)
ndtype = _check_object_for_strings(values)
htable = _hashtables.get(ndtype, _hashtables["object"])
return htable, values
def _check_object_for_strings(values: np.ndarray) -> str:
"""
Check if we can use string hashtable instead of object hashtable.
Parameters
----------
values : ndarray
Returns
-------
str
"""
ndtype = values.dtype.name
if ndtype == "object":
# it's cheaper to use a String Hash Table than Object; we infer
# including nulls because that is the only difference between
# StringHashTable and ObjectHashtable
if lib.infer_dtype(values, skipna=False) in ["string"]:
ndtype = "string"
return ndtype
# --------------- #
# top-level algos #
# --------------- #
def unique(values):
"""
Hash table-based unique. Uniques are returned in order
of appearance. This does NOT sort.
Significantly faster than numpy.unique for long enough sequences.
Includes NA values.
Parameters
----------
values : 1d array-like
Returns
-------
numpy.ndarray or ExtensionArray
The return can be:
* Index : when the input is an Index
* Categorical : when the input is a Categorical dtype
* ndarray : when the input is a Series/ndarray
Return numpy.ndarray or ExtensionArray.
See Also
--------
Index.unique : Return unique values from an Index.
Series.unique : Return unique values of Series object.
Examples
--------
>>> pd.unique(pd.Series([2, 1, 3, 3]))
array([2, 1, 3])
>>> pd.unique(pd.Series([2] + [1] * 5))
array([2, 1])
>>> pd.unique(pd.Series([pd.Timestamp("20160101"), pd.Timestamp("20160101")]))
array(['2016-01-01T00:00:00.000000000'], dtype='datetime64[ns]')
>>> pd.unique(
... pd.Series(
... [
... pd.Timestamp("20160101", tz="US/Eastern"),
... pd.Timestamp("20160101", tz="US/Eastern"),
... ]
... )
... )
<DatetimeArray>
['2016-01-01 00:00:00-05:00']
Length: 1, dtype: datetime64[ns, US/Eastern]
>>> pd.unique(
... pd.Index(
... [
... pd.Timestamp("20160101", tz="US/Eastern"),
... pd.Timestamp("20160101", tz="US/Eastern"),
... ]
... )
... )
DatetimeIndex(['2016-01-01 00:00:00-05:00'],
dtype='datetime64[ns, US/Eastern]',
freq=None)
>>> pd.unique(list("baabc"))
array(['b', 'a', 'c'], dtype=object)
An unordered Categorical will return categories in the
order of appearance.
>>> pd.unique(pd.Series(pd.Categorical(list("baabc"))))
['b', 'a', 'c']
Categories (3, object): ['a', 'b', 'c']
>>> pd.unique(pd.Series(pd.Categorical(list("baabc"), categories=list("abc"))))
['b', 'a', 'c']
Categories (3, object): ['a', 'b', 'c']
An ordered Categorical preserves the category ordering.
>>> pd.unique(
... pd.Series(
... pd.Categorical(list("baabc"), categories=list("abc"), ordered=True)
... )
... )
['b', 'a', 'c']
Categories (3, object): ['a' < 'b' < 'c']
An array of tuples
>>> pd.unique([("a", "b"), ("b", "a"), ("a", "c"), ("b", "a")])
array([('a', 'b'), ('b', 'a'), ('a', 'c')], dtype=object)
"""
values = _ensure_arraylike(values)
if is_extension_array_dtype(values.dtype):
# Dispatch to extension dtype's unique.
return values.unique()
original = values
htable, values = _get_hashtable_algo(values)
table = htable(len(values))
uniques = table.unique(values)
uniques = _reconstruct_data(uniques, original.dtype, original)
return uniques
unique1d = unique
def isin(comps: AnyArrayLike, values: AnyArrayLike) -> npt.NDArray[np.bool_]:
"""
Compute the isin boolean array.
Parameters
----------
comps : array-like
values : array-like
Returns
-------
ndarray[bool]
Same length as `comps`.
"""
if not is_list_like(comps):
raise TypeError(
"only list-like objects are allowed to be passed "
f"to isin(), you passed a [{type(comps).__name__}]"
)
if not is_list_like(values):
raise TypeError(
"only list-like objects are allowed to be passed "
f"to isin(), you passed a [{type(values).__name__}]"
)
if not isinstance(values, (ABCIndex, ABCSeries, ABCExtensionArray, np.ndarray)):
values = _ensure_arraylike(list(values))
elif isinstance(values, ABCMultiIndex):
# Avoid raising in extract_array
values = np.array(values)
else:
values = extract_array(values, extract_numpy=True, extract_range=True)
comps = _ensure_arraylike(comps)
comps = extract_array(comps, extract_numpy=True)
if not isinstance(comps, np.ndarray):
# i.e. Extension Array
return comps.isin(values)
elif needs_i8_conversion(comps.dtype):
# Dispatch to DatetimeLikeArrayMixin.isin
return pd_array(comps).isin(values)
elif needs_i8_conversion(values.dtype) and not is_object_dtype(comps.dtype):
# e.g. comps are integers and values are datetime64s
return np.zeros(comps.shape, dtype=bool)
# TODO: not quite right ... Sparse/Categorical
elif needs_i8_conversion(values.dtype):
return isin(comps, values.astype(object))
elif is_extension_array_dtype(values.dtype):
return isin(np.asarray(comps), np.asarray(values))
# GH16012
# Ensure np.in1d doesn't get object types or it *may* throw an exception
# Albeit hashmap has O(1) look-up (vs. O(logn) in sorted array),
# in1d is faster for small sizes
if len(comps) > 1_000_000 and len(values) <= 26 and not is_object_dtype(comps):
# If the values include nan we need to check for nan explicitly
# since np.nan it not equal to np.nan
if isna(values).any():
def f(c, v):
return np.logical_or(np.in1d(c, v), np.isnan(c))
else:
f = np.in1d
else:
# error: List item 0 has incompatible type "Union[Any, dtype[Any],
# ExtensionDtype]"; expected "Union[dtype[Any], None, type, _SupportsDType, str,
# Tuple[Any, Union[int, Sequence[int]]], List[Any], _DTypeDict, Tuple[Any,
# Any]]"
# error: List item 1 has incompatible type "Union[Any, ExtensionDtype]";
# expected "Union[dtype[Any], None, type, _SupportsDType, str, Tuple[Any,
# Union[int, Sequence[int]]], List[Any], _DTypeDict, Tuple[Any, Any]]"
# error: List item 1 has incompatible type "Union[dtype[Any], ExtensionDtype]";
# expected "Union[dtype[Any], None, type, _SupportsDType, str, Tuple[Any,
# Union[int, Sequence[int]]], List[Any], _DTypeDict, Tuple[Any, Any]]"
common = np.find_common_type(
[values.dtype, comps.dtype], [] # type: ignore[list-item]
)
values = values.astype(common, copy=False)
comps = comps.astype(common, copy=False)
f = htable.ismember
return f(comps, values)
def factorize_array(
values: np.ndarray,
na_sentinel: int = -1,
size_hint: int | None = None,
na_value=None,
mask: np.ndarray | None = None,
) -> tuple[npt.NDArray[np.intp], np.ndarray]:
"""
Factorize a numpy array to codes and uniques.
This doesn't do any coercion of types or unboxing before factorization.
Parameters
----------
values : ndarray
na_sentinel : int, default -1
size_hint : int, optional
Passed through to the hashtable's 'get_labels' method
na_value : object, optional
A value in `values` to consider missing. Note: only use this
parameter when you know that you don't have any values pandas would
consider missing in the array (NaN for float data, iNaT for
datetimes, etc.).
mask : ndarray[bool], optional
If not None, the mask is used as indicator for missing values
(True = missing, False = valid) instead of `na_value` or
condition "val != val".
Returns
-------
codes : ndarray[np.intp]
uniques : ndarray
"""
hash_klass, values = get_data_algo(values)
table = hash_klass(size_hint or len(values))
uniques, codes = table.factorize(
values, na_sentinel=na_sentinel, na_value=na_value, mask=mask
)
codes = ensure_platform_int(codes)
return codes, uniques
@doc(
values=dedent(
"""\
values : sequence
A 1-D sequence. Sequences that aren't pandas objects are
coerced to ndarrays before factorization.
"""
),
sort=dedent(
"""\
sort : bool, default False
Sort `uniques` and shuffle `codes` to maintain the
relationship.
"""
),
size_hint=dedent(
"""\
size_hint : int, optional
Hint to the hashtable sizer.
"""
),
)
def factorize(
values,
sort: bool = False,
na_sentinel: int | None = -1,
size_hint: int | None = None,
) -> tuple[np.ndarray, np.ndarray | Index]:
"""
Encode the object as an enumerated type or categorical variable.
This method is useful for obtaining a numeric representation of an
array when all that matters is identifying distinct values. `factorize`
is available as both a top-level function :func:`pandas.factorize`,
and as a method :meth:`Series.factorize` and :meth:`Index.factorize`.
Parameters
----------
{values}{sort}
na_sentinel : int or None, default -1
Value to mark "not found". If None, will not drop the NaN
from the uniques of the values.
.. versionchanged:: 1.1.2
{size_hint}\
Returns
-------
codes : ndarray
An integer ndarray that's an indexer into `uniques`.
``uniques.take(codes)`` will have the same values as `values`.
uniques : ndarray, Index, or Categorical
The unique valid values. When `values` is Categorical, `uniques`
is a Categorical. When `values` is some other pandas object, an
`Index` is returned. Otherwise, a 1-D ndarray is returned.
.. note ::
Even if there's a missing value in `values`, `uniques` will
*not* contain an entry for it.
See Also
--------
cut : Discretize continuous-valued array.
unique : Find the unique value in an array.
Examples
--------
These examples all show factorize as a top-level method like
``pd.factorize(values)``. The results are identical for methods like
:meth:`Series.factorize`.
>>> codes, uniques = pd.factorize(['b', 'b', 'a', 'c', 'b'])
>>> codes
array([0, 0, 1, 2, 0]...)
>>> uniques
array(['b', 'a', 'c'], dtype=object)
With ``sort=True``, the `uniques` will be sorted, and `codes` will be
shuffled so that the relationship is the maintained.
>>> codes, uniques = pd.factorize(['b', 'b', 'a', 'c', 'b'], sort=True)
>>> codes
array([1, 1, 0, 2, 1]...)
>>> uniques
array(['a', 'b', 'c'], dtype=object)
Missing values are indicated in `codes` with `na_sentinel`
(``-1`` by default). Note that missing values are never
included in `uniques`.
>>> codes, uniques = pd.factorize(['b', None, 'a', 'c', 'b'])
>>> codes
array([ 0, -1, 1, 2, 0]...)
>>> uniques
array(['b', 'a', 'c'], dtype=object)
Thus far, we've only factorized lists (which are internally coerced to
NumPy arrays). When factorizing pandas objects, the type of `uniques`
will differ. For Categoricals, a `Categorical` is returned.
>>> cat = pd.Categorical(['a', 'a', 'c'], categories=['a', 'b', 'c'])
>>> codes, uniques = pd.factorize(cat)
>>> codes
array([0, 0, 1]...)
>>> uniques
['a', 'c']
Categories (3, object): ['a', 'b', 'c']
Notice that ``'b'`` is in ``uniques.categories``, despite not being
present in ``cat.values``.
For all other pandas objects, an Index of the appropriate type is
returned.
>>> cat = pd.Series(['a', 'a', 'c'])
>>> codes, uniques = pd.factorize(cat)
>>> codes
array([0, 0, 1]...)
>>> uniques
Index(['a', 'c'], dtype='object')
If NaN is in the values, and we want to include NaN in the uniques of the
values, it can be achieved by setting ``na_sentinel=None``.
>>> values = np.array([1, 2, 1, np.nan])
>>> codes, uniques = pd.factorize(values) # default: na_sentinel=-1
>>> codes
array([ 0, 1, 0, -1])
>>> uniques
array([1., 2.])
>>> codes, uniques = pd.factorize(values, na_sentinel=None)
>>> codes
array([0, 1, 0, 2])
>>> uniques
array([ 1., 2., nan])
"""
# Implementation notes: This method is responsible for 3 things
# 1.) coercing data to array-like (ndarray, Index, extension array)
# 2.) factorizing codes and uniques
# 3.) Maybe boxing the uniques in an Index
#
# Step 2 is dispatched to extension types (like Categorical). They are
# responsible only for factorization. All data coercion, sorting and boxing
# should happen here.
if isinstance(values, ABCRangeIndex):
return values.factorize(sort=sort)
values = _ensure_arraylike(values)
original = values
if not isinstance(values, ABCMultiIndex):
values = extract_array(values, extract_numpy=True)
# GH35667, if na_sentinel=None, we will not dropna NaNs from the uniques
# of values, assign na_sentinel=-1 to replace code value for NaN.
dropna = True
if na_sentinel is None:
na_sentinel = -1
dropna = False
if (
isinstance(values, (ABCDatetimeArray, ABCTimedeltaArray))
and values.freq is not None
):
codes, uniques = values.factorize(sort=sort)
if isinstance(original, ABCIndex):
uniques = original._shallow_copy(uniques, name=None)
elif isinstance(original, ABCSeries):
from pandas import Index
uniques = Index(uniques)
return codes, uniques
if not isinstance(values.dtype, np.dtype):
# i.e. ExtensionDtype
codes, uniques = values.factorize(na_sentinel=na_sentinel)
dtype = original.dtype
else:
dtype = values.dtype
values = _ensure_data(values)
na_value: Scalar
if original.dtype.kind in ["m", "M"]:
# Note: factorize_array will cast NaT bc it has a __int__
# method, but will not cast the more-correct dtype.type("nat")
na_value = iNaT
else:
na_value = None
codes, uniques = factorize_array(
values, na_sentinel=na_sentinel, size_hint=size_hint, na_value=na_value
)
if sort and len(uniques) > 0:
uniques, codes = safe_sort(
uniques, codes, na_sentinel=na_sentinel, assume_unique=True, verify=False
)
code_is_na = codes == na_sentinel
if not dropna and code_is_na.any():
# na_value is set based on the dtype of uniques, and compat set to False is
# because we do not want na_value to be 0 for integers
na_value = na_value_for_dtype(uniques.dtype, compat=False)
uniques = np.append(uniques, [na_value])
codes = np.where(code_is_na, len(uniques) - 1, codes)
uniques = _reconstruct_data(uniques, dtype, original)
# return original tenor
if isinstance(original, ABCIndex):
if original.dtype.kind in ["m", "M"] and isinstance(uniques, np.ndarray):
original._data = cast(
"Union[DatetimeArray, TimedeltaArray]", original._data
)
uniques = type(original._data)._simple_new(uniques, dtype=original.dtype)
uniques = original._shallow_copy(uniques, name=None)
elif isinstance(original, ABCSeries):
from pandas import Index
uniques = Index(uniques)
return codes, uniques
def value_counts(
values,
sort: bool = True,
ascending: bool = False,
normalize: bool = False,
bins=None,
dropna: bool = True,
) -> Series:
"""
Compute a histogram of the counts of non-null values.
Parameters
----------
values : ndarray (1-d)
sort : bool, default True
Sort by values
ascending : bool, default False
Sort in ascending order
normalize: bool, default False
If True then compute a relative histogram
bins : integer, optional
Rather than count values, group them into half-open bins,
convenience for pd.cut, only works with numeric data
dropna : bool, default True
Don't include counts of NaN
Returns
-------
Series
"""
from pandas.core.series import Series
name = getattr(values, "name", None)
if bins is not None:
from pandas.core.reshape.tile import cut
values = Series(values)
try:
ii = cut(values, bins, include_lowest=True)
except TypeError as err:
raise TypeError("bins argument only works with numeric data.") from err
# count, remove nulls (from the index), and but the bins
result = ii.value_counts(dropna=dropna)
result = result[result.index.notna()]
result.index = result.index.astype("interval")
result = result.sort_index()
# if we are dropna and we have NO values
if dropna and (result._values == 0).all():
result = result.iloc[0:0]
# normalizing is by len of all (regardless of dropna)
counts = np.array([len(ii)])
else:
if is_extension_array_dtype(values):
# handle Categorical and sparse,
result = Series(values)._values.value_counts(dropna=dropna)
result.name = name
counts = result._values
else:
keys, counts = value_counts_arraylike(values, dropna)
result = Series(counts, index=keys, name=name)
if sort:
result = result.sort_values(ascending=ascending)
if normalize:
result = result / counts.sum()
return result
# Called once from SparseArray, otherwise could be private
def value_counts_arraylike(values, dropna: bool):
"""
Parameters
----------
values : arraylike
dropna : bool
Returns
-------
uniques : np.ndarray or ExtensionArray
counts : np.ndarray
"""
values = _ensure_arraylike(values)
original = values
values = _ensure_data(values)
# TODO: handle uint8
keys, counts = htable.value_count(values, dropna)
if needs_i8_conversion(original.dtype):
# datetime, timedelta, or period
if dropna:
msk = keys != iNaT
keys, counts = keys[msk], counts[msk]
res_keys = _reconstruct_data(keys, original.dtype, original)
return res_keys, counts
def duplicated(
values: ArrayLike, keep: Literal["first", "last", False] = "first"
) -> npt.NDArray[np.bool_]:
"""
Return boolean ndarray denoting duplicate values.
Parameters
----------
values : nd.array, ExtensionArray or Series
Array over which to check for duplicate values.
keep : {'first', 'last', False}, default 'first'
- ``first`` : Mark duplicates as ``True`` except for the first
occurrence.
- ``last`` : Mark duplicates as ``True`` except for the last
occurrence.
- False : Mark all duplicates as ``True``.
Returns
-------
duplicated : ndarray[bool]
"""
values = _ensure_data(values)
return htable.duplicated(values, keep=keep)
def mode(values, dropna: bool = True) -> Series:
"""
Returns the mode(s) of an array.
Parameters
----------
values : array-like
Array over which to check for duplicate values.
dropna : bool, default True
Don't consider counts of NaN/NaT.
Returns
-------
mode : Series
"""
from pandas import Series
from pandas.core.indexes.api import default_index
values = _ensure_arraylike(values)
original = values
# categorical is a fast-path
if is_categorical_dtype(values):
if isinstance(values, Series):
# TODO: should we be passing `name` below?
return Series(values._values.mode(dropna=dropna), name=values.name)
return values.mode(dropna=dropna)
if dropna and needs_i8_conversion(values.dtype):
mask = values.isnull()
values = values[~mask]
values = _ensure_data(values)
npresult = htable.mode(values, dropna=dropna)
try:
npresult = np.sort(npresult)
except TypeError as err:
warn(f"Unable to sort modes: {err}")
result = _reconstruct_data(npresult, original.dtype, original)
# Ensure index is type stable (should always use int index)
return Series(result, index=default_index(len(result)))
def rank(
values: ArrayLike,
axis: int = 0,
method: str = "average",
na_option: str = "keep",
ascending: bool = True,
pct: bool = False,
) -> np.ndarray:
"""
Rank the values along a given axis.
Parameters
----------
values : array-like
Array whose values will be ranked. The number of dimensions in this
array must not exceed 2.
axis : int, default 0
Axis over which to perform rankings.
method : {'average', 'min', 'max', 'first', 'dense'}, default 'average'
The method by which tiebreaks are broken during the ranking.
na_option : {'keep', 'top'}, default 'keep'
The method by which NaNs are placed in the ranking.
- ``keep``: rank each NaN value with a NaN ranking
- ``top``: replace each NaN with either +/- inf so that they
there are ranked at the top
ascending : bool, default True
Whether or not the elements should be ranked in ascending order.
pct : bool, default False
Whether or not to the display the returned rankings in integer form
(e.g. 1, 2, 3) or in percentile form (e.g. 0.333..., 0.666..., 1).
"""
is_datetimelike = needs_i8_conversion(values.dtype)
values = _get_values_for_rank(values)
if values.ndim == 1:
ranks = algos.rank_1d(
values,
is_datetimelike=is_datetimelike,
ties_method=method,
ascending=ascending,
na_option=na_option,
pct=pct,
)
elif values.ndim == 2:
ranks = algos.rank_2d(
values,
axis=axis,
is_datetimelike=is_datetimelike,
ties_method=method,
ascending=ascending,
na_option=na_option,
pct=pct,
)
else:
raise TypeError("Array with ndim > 2 are not supported.")
return ranks
def checked_add_with_arr(
arr: np.ndarray,
b,
arr_mask: npt.NDArray[np.bool_] | None = None,
b_mask: npt.NDArray[np.bool_] | None = None,
) -> np.ndarray:
"""
Perform array addition that checks for underflow and overflow.
Performs the addition of an int64 array and an int64 integer (or array)
but checks that they do not result in overflow first. For elements that
are indicated to be NaN, whether or not there is overflow for that element
is automatically ignored.
Parameters
----------
arr : array addend.
b : array or scalar addend.
arr_mask : np.ndarray[bool] or None, default None
array indicating which elements to exclude from checking
b_mask : np.ndarray[bool] or None, default None
array or scalar indicating which element(s) to exclude from checking
Returns
-------
sum : An array for elements x + b for each element x in arr if b is
a scalar or an array for elements x + y for each element pair
(x, y) in (arr, b).
Raises
------
OverflowError if any x + y exceeds the maximum or minimum int64 value.
"""
# For performance reasons, we broadcast 'b' to the new array 'b2'
# so that it has the same size as 'arr'.
b2 = np.broadcast_to(b, arr.shape)
if b_mask is not None:
# We do the same broadcasting for b_mask as well.
b2_mask = np.broadcast_to(b_mask, arr.shape)
else:
b2_mask = None
# For elements that are NaN, regardless of their value, we should
# ignore whether they overflow or not when doing the checked add.
if arr_mask is not None and b2_mask is not None:
not_nan = np.logical_not(arr_mask | b2_mask)
elif arr_mask is not None:
not_nan = np.logical_not(arr_mask)
elif b_mask is not None:
not_nan = np.logical_not(b2_mask)
else:
not_nan = np.empty(arr.shape, dtype=bool)
not_nan.fill(True)
# gh-14324: For each element in 'arr' and its corresponding element
# in 'b2', we check the sign of the element in 'b2'. If it is positive,
# we then check whether its sum with the element in 'arr' exceeds
# np.iinfo(np.int64).max. If so, we have an overflow error. If it
# it is negative, we then check whether its sum with the element in
# 'arr' exceeds np.iinfo(np.int64).min. If so, we have an overflow
# error as well.
i8max = lib.i8max
i8min = iNaT
mask1 = b2 > 0
mask2 = b2 < 0
if not mask1.any():
to_raise = ((i8min - b2 > arr) & not_nan).any()
elif not mask2.any():
to_raise = ((i8max - b2 < arr) & not_nan).any()
else:
to_raise = ((i8max - b2[mask1] < arr[mask1]) & not_nan[mask1]).any() or (
(i8min - b2[mask2] > arr[mask2]) & not_nan[mask2]
).any()
if to_raise:
raise OverflowError("Overflow in int64 addition")
return arr + b
def quantile(x, q, interpolation_method="fraction"):
"""
Compute sample quantile or quantiles of the input array. For example, q=0.5
computes the median.
The `interpolation_method` parameter supports three values, namely
`fraction` (default), `lower` and `higher`. Interpolation is done only,
if the desired quantile lies between two data points `i` and `j`. For
`fraction`, the result is an interpolated value between `i` and `j`;
for `lower`, the result is `i`, for `higher` the result is `j`.
Parameters
----------
x : ndarray
Values from which to extract score.
q : scalar or array
Percentile at which to extract score.
interpolation_method : {'fraction', 'lower', 'higher'}, optional
This optional parameter specifies the interpolation method to use,
when the desired quantile lies between two data points `i` and `j`:
- fraction: `i + (j - i)*fraction`, where `fraction` is the
fractional part of the index surrounded by `i` and `j`.
-lower: `i`.
- higher: `j`.
Returns
-------
score : float
Score at percentile.
Examples
--------
>>> from scipy import stats
>>> a = np.arange(100)
>>> stats.scoreatpercentile(a, 50)
49.5
"""
x = np.asarray(x)
mask = isna(x)
x = x[~mask]
values = np.sort(x)
def _interpolate(a, b, fraction):
"""
Returns the point at the given fraction between a and b, where
'fraction' must be between 0 and 1.
"""
return a + (b - a) * fraction
def _get_score(at):
if len(values) == 0:
return np.nan
idx = at * (len(values) - 1)
if idx % 1 == 0:
score = values[int(idx)]
else:
if interpolation_method == "fraction":
score = _interpolate(values[int(idx)], values[int(idx) + 1], idx % 1)
elif interpolation_method == "lower":
score = values[np.floor(idx)]
elif interpolation_method == "higher":
score = values[np.ceil(idx)]
else:
raise ValueError(
"interpolation_method can only be 'fraction' "
", 'lower' or 'higher'"
)
return score
if is_scalar(q):
return _get_score(q)
q = np.asarray(q, np.float64)
result = [_get_score(x) for x in q]
return np.array(result, dtype=np.float64)
# --------------- #
# select n #
# --------------- #
class SelectN:
def __init__(self, obj, n: int, keep: str):
self.obj = obj
self.n = n
self.keep = keep
if self.keep not in ("first", "last", "all"):
raise ValueError('keep must be either "first", "last" or "all"')
def compute(self, method: str) -> DataFrame | Series:
raise NotImplementedError
@final
def nlargest(self):
return self.compute("nlargest")
@final
def nsmallest(self):
return self.compute("nsmallest")
@final
@staticmethod
def is_valid_dtype_n_method(dtype: DtypeObj) -> bool:
"""
Helper function to determine if dtype is valid for
nsmallest/nlargest methods
"""
return (
is_numeric_dtype(dtype) and not is_complex_dtype(dtype)
) or needs_i8_conversion(dtype)
class SelectNSeries(SelectN):
"""
Implement n largest/smallest for Series
Parameters
----------
obj : Series
n : int
keep : {'first', 'last'}, default 'first'
Returns
-------
nordered : Series
"""
def compute(self, method: str) -> Series:
from pandas.core.reshape.concat import concat
n = self.n
dtype = self.obj.dtype
if not self.is_valid_dtype_n_method(dtype):
raise TypeError(f"Cannot use method '{method}' with dtype {dtype}")
if n <= 0:
return self.obj[[]]
dropped = self.obj.dropna()
nan_index = self.obj.drop(dropped.index)
if is_extension_array_dtype(dropped.dtype):
# GH#41816 bc we have dropped NAs above, MaskedArrays can use the
# numpy logic.
from pandas.core.arrays import BaseMaskedArray
arr = dropped._values
if isinstance(arr, BaseMaskedArray):
ser = type(dropped)(arr._data, index=dropped.index, name=dropped.name)
result = type(self)(ser, n=self.n, keep=self.keep).compute(method)
return result.astype(arr.dtype)
# slow method
if n >= len(self.obj):
ascending = method == "nsmallest"
return self.obj.sort_values(ascending=ascending).head(n)
# fast method
new_dtype = dropped.dtype
arr = _ensure_data(dropped.values)
if method == "nlargest":
arr = -arr
if is_integer_dtype(new_dtype):
# GH 21426: ensure reverse ordering at boundaries
arr -= 1
elif is_bool_dtype(new_dtype):
# GH 26154: ensure False is smaller than True
arr = 1 - (-arr)
if self.keep == "last":
arr = arr[::-1]
nbase = n
findex = len(self.obj)
narr = len(arr)
n = min(n, narr)
# arr passed into kth_smallest must be contiguous. We copy
# here because kth_smallest will modify its input
kth_val = algos.kth_smallest(arr.copy(order="C"), n - 1)
(ns,) = np.nonzero(arr <= kth_val)
inds = ns[arr[ns].argsort(kind="mergesort")]
if self.keep != "all":
inds = inds[:n]
findex = nbase
if self.keep == "last":
# reverse indices
inds = narr - 1 - inds
return concat([dropped.iloc[inds], nan_index]).iloc[:findex]
class SelectNFrame(SelectN):
"""
Implement n largest/smallest for DataFrame
Parameters
----------
obj : DataFrame
n : int
keep : {'first', 'last'}, default 'first'
columns : list or str
Returns
-------
nordered : DataFrame
"""
def __init__(self, obj, n: int, keep: str, columns):
super().__init__(obj, n, keep)
if not is_list_like(columns) or isinstance(columns, tuple):
columns = [columns]
columns = list(columns)
self.columns = columns
def compute(self, method: str) -> DataFrame:
from pandas.core.api import Int64Index
n = self.n
frame = self.obj
columns = self.columns
for column in columns:
dtype = frame[column].dtype
if not self.is_valid_dtype_n_method(dtype):
raise TypeError(
f"Column {repr(column)} has dtype {dtype}, "
f"cannot use method {repr(method)} with this dtype"
)
def get_indexer(current_indexer, other_indexer):
"""
Helper function to concat `current_indexer` and `other_indexer`
depending on `method`
"""
if method == "nsmallest":
return current_indexer.append(other_indexer)
else:
return other_indexer.append(current_indexer)
# Below we save and reset the index in case index contains duplicates
original_index = frame.index
cur_frame = frame = frame.reset_index(drop=True)
cur_n = n
indexer = Int64Index([])
for i, column in enumerate(columns):
# For each column we apply method to cur_frame[column].
# If it's the last column or if we have the number of
# results desired we are done.
# Otherwise there are duplicates of the largest/smallest
# value and we need to look at the rest of the columns
# to determine which of the rows with the largest/smallest
# value in the column to keep.
series = cur_frame[column]
is_last_column = len(columns) - 1 == i
values = getattr(series, method)(
cur_n, keep=self.keep if is_last_column else "all"
)
if is_last_column or len(values) <= cur_n:
indexer = get_indexer(indexer, values.index)
break
# Now find all values which are equal to
# the (nsmallest: largest)/(nlargest: smallest)
# from our series.
border_value = values == values[values.index[-1]]
# Some of these values are among the top-n
# some aren't.
unsafe_values = values[border_value]
# These values are definitely among the top-n
safe_values = values[~border_value]
indexer = get_indexer(indexer, safe_values.index)
# Go on and separate the unsafe_values on the remaining
# columns.
cur_frame = cur_frame.loc[unsafe_values.index]
cur_n = n - len(indexer)
frame = frame.take(indexer)
# Restore the index on frame
frame.index = original_index.take(indexer)
# If there is only one column, the frame is already sorted.
if len(columns) == 1:
return frame
ascending = method == "nsmallest"
return frame.sort_values(columns, ascending=ascending, kind="mergesort")
# ---- #
# take #
# ---- #
def take(
arr,
indices: TakeIndexer,
axis: int = 0,
allow_fill: bool = False,
fill_value=None,
):
"""
Take elements from an array.
Parameters
----------
arr : array-like or scalar value
Non array-likes (sequences/scalars without a dtype) are coerced
to an ndarray.
indices : sequence of int or one-dimensional np.ndarray of int
Indices to be taken.
axis : int, default 0
The axis over which to select values.
allow_fill : bool, default False
How to handle negative values in `indices`.
* False: negative values in `indices` indicate positional indices
from the right (the default). This is similar to :func:`numpy.take`.
* True: negative values in `indices` indicate
missing values. These values are set to `fill_value`. Any other
negative values raise a ``ValueError``.
fill_value : any, optional
Fill value to use for NA-indices when `allow_fill` is True.
This may be ``None``, in which case the default NA value for
the type (``self.dtype.na_value``) is used.
For multi-dimensional `arr`, each *element* is filled with
`fill_value`.
Returns
-------
ndarray or ExtensionArray
Same type as the input.
Raises
------
IndexError
When `indices` is out of bounds for the array.
ValueError
When the indexer contains negative values other than ``-1``
and `allow_fill` is True.
Notes
-----
When `allow_fill` is False, `indices` may be whatever dimensionality
is accepted by NumPy for `arr`.
When `allow_fill` is True, `indices` should be 1-D.
See Also
--------
numpy.take : Take elements from an array along an axis.
Examples
--------
>>> from pandas.api.extensions import take
With the default ``allow_fill=False``, negative numbers indicate
positional indices from the right.
>>> take(np.array([10, 20, 30]), [0, 0, -1])
array([10, 10, 30])
Setting ``allow_fill=True`` will place `fill_value` in those positions.
>>> take(np.array([10, 20, 30]), [0, 0, -1], allow_fill=True)
array([10., 10., nan])
>>> take(np.array([10, 20, 30]), [0, 0, -1], allow_fill=True,
... fill_value=-10)
array([ 10, 10, -10])
"""
if not is_array_like(arr):
arr = np.asarray(arr)
indices = np.asarray(indices, dtype=np.intp)
if allow_fill:
# Pandas style, -1 means NA
validate_indices(indices, arr.shape[axis])
result = take_nd(
arr, indices, axis=axis, allow_fill=True, fill_value=fill_value
)
else:
# NumPy style
result = arr.take(indices, axis=axis)
return result
# ------------ #
# searchsorted #
# ------------ #
def searchsorted(
arr: ArrayLike,
value: NumpyValueArrayLike | ExtensionArray,
side: Literal["left", "right"] = "left",
sorter: NumpySorter = None,
) -> npt.NDArray[np.intp] | np.intp:
"""
Find indices where elements should be inserted to maintain order.
.. versionadded:: 0.25.0
Find the indices into a sorted array `arr` (a) such that, if the
corresponding elements in `value` were inserted before the indices,
the order of `arr` would be preserved.
Assuming that `arr` is sorted:
====== ================================
`side` returned index `i` satisfies
====== ================================
left ``arr[i-1] < value <= self[i]``
right ``arr[i-1] <= value < self[i]``
====== ================================
Parameters
----------
arr: np.ndarray, ExtensionArray, Series
Input array. If `sorter` is None, then it must be sorted in
ascending order, otherwise `sorter` must be an array of indices
that sort it.
value : array-like or scalar
Values to insert into `arr`.
side : {'left', 'right'}, optional
If 'left', the index of the first suitable location found is given.
If 'right', return the last such index. If there is no suitable
index, return either 0 or N (where N is the length of `self`).
sorter : 1-D array-like, optional
Optional array of integer indices that sort array a into ascending
order. They are typically the result of argsort.
Returns
-------
array of ints or int
If value is array-like, array of insertion points.
If value is scalar, a single integer.
See Also
--------
numpy.searchsorted : Similar method from NumPy.
"""
if sorter is not None:
sorter = ensure_platform_int(sorter)
if (
isinstance(arr, np.ndarray)
and is_integer_dtype(arr.dtype)
and (is_integer(value) or is_integer_dtype(value))
):
# if `arr` and `value` have different dtypes, `arr` would be
# recast by numpy, causing a slow search.
# Before searching below, we therefore try to give `value` the
# same dtype as `arr`, while guarding against integer overflows.
iinfo = np.iinfo(arr.dtype.type)
value_arr = np.array([value]) if is_scalar(value) else np.array(value)
if (value_arr >= iinfo.min).all() and (value_arr <= iinfo.max).all():
# value within bounds, so no overflow, so can convert value dtype
# to dtype of arr
dtype = arr.dtype
else:
dtype = value_arr.dtype
if is_scalar(value):
# We know that value is int
value = cast(int, dtype.type(value))
else:
value = pd_array(cast(ArrayLike, value), dtype=dtype)
elif not (
is_object_dtype(arr) or is_numeric_dtype(arr) or is_categorical_dtype(arr)
):
# E.g. if `arr` is an array with dtype='datetime64[ns]'
# and `value` is a pd.Timestamp, we may need to convert value
arr = ensure_wrapped_if_datetimelike(arr)
# Argument 1 to "searchsorted" of "ndarray" has incompatible type
# "Union[NumpyValueArrayLike, ExtensionArray]"; expected "NumpyValueArrayLike"
return arr.searchsorted(value, side=side, sorter=sorter) # type: ignore[arg-type]
# ---- #
# diff #
# ---- #
_diff_special = {"float64", "float32", "int64", "int32", "int16", "int8"}
def diff(arr, n: int, axis: int = 0, stacklevel: int = 3):
"""
difference of n between self,
analogous to s-s.shift(n)
Parameters
----------
arr : ndarray or ExtensionArray
n : int
number of periods
axis : {0, 1}
axis to shift on
stacklevel : int, default 3
The stacklevel for the lost dtype warning.
Returns
-------
shifted
"""
n = int(n)
na = np.nan
dtype = arr.dtype
is_bool = is_bool_dtype(dtype)
if is_bool:
op = operator.xor
else:
op = operator.sub
if isinstance(dtype, PandasDtype):
# PandasArray cannot necessarily hold shifted versions of itself.
arr = arr.to_numpy()
dtype = arr.dtype
if not isinstance(dtype, np.dtype):
# i.e ExtensionDtype
if hasattr(arr, f"__{op.__name__}__"):
if axis != 0:
raise ValueError(f"cannot diff {type(arr).__name__} on axis={axis}")
return op(arr, arr.shift(n))
else:
warn(
"dtype lost in 'diff()'. In the future this will raise a "
"TypeError. Convert to a suitable dtype prior to calling 'diff'.",
FutureWarning,
stacklevel=stacklevel,
)
arr = np.asarray(arr)
dtype = arr.dtype
is_timedelta = False
if needs_i8_conversion(arr.dtype):
dtype = np.int64
arr = arr.view("i8")
na = iNaT
is_timedelta = True
elif is_bool:
# We have to cast in order to be able to hold np.nan
dtype = np.object_
elif is_integer_dtype(dtype):
# We have to cast in order to be able to hold np.nan
# int8, int16 are incompatible with float64,
# see https://github.com/cython/cython/issues/2646
if arr.dtype.name in ["int8", "int16"]:
dtype = np.float32
else:
dtype = np.float64
orig_ndim = arr.ndim
if orig_ndim == 1:
# reshape so we can always use algos.diff_2d
arr = arr.reshape(-1, 1)
# TODO: require axis == 0
dtype = np.dtype(dtype)
out_arr = np.empty(arr.shape, dtype=dtype)
na_indexer = [slice(None)] * 2
na_indexer[axis] = slice(None, n) if n >= 0 else slice(n, None)
out_arr[tuple(na_indexer)] = na
if arr.dtype.name in _diff_special:
# TODO: can diff_2d dtype specialization troubles be fixed by defining
# out_arr inside diff_2d?
algos.diff_2d(arr, out_arr, n, axis, datetimelike=is_timedelta)
else:
# To keep mypy happy, _res_indexer is a list while res_indexer is
# a tuple, ditto for lag_indexer.
_res_indexer = [slice(None)] * 2
_res_indexer[axis] = slice(n, None) if n >= 0 else slice(None, n)
res_indexer = tuple(_res_indexer)
_lag_indexer = [slice(None)] * 2
_lag_indexer[axis] = slice(None, -n) if n > 0 else slice(-n, None)
lag_indexer = tuple(_lag_indexer)
out_arr[res_indexer] = op(arr[res_indexer], arr[lag_indexer])
if is_timedelta:
out_arr = out_arr.view("timedelta64[ns]")
if orig_ndim == 1:
out_arr = out_arr[:, 0]
return out_arr
# --------------------------------------------------------------------
# Helper functions
# Note: safe_sort is in algorithms.py instead of sorting.py because it is
# low-dependency, is used in this module, and used private methods from
# this module.
def safe_sort(
values,
codes=None,
na_sentinel: int = -1,
assume_unique: bool = False,
verify: bool = True,
) -> np.ndarray | tuple[np.ndarray, np.ndarray]:
"""
Sort ``values`` and reorder corresponding ``codes``.
``values`` should be unique if ``codes`` is not None.
Safe for use with mixed types (int, str), orders ints before strs.
Parameters
----------
values : list-like
Sequence; must be unique if ``codes`` is not None.
codes : list_like, optional
Indices to ``values``. All out of bound indices are treated as
"not found" and will be masked with ``na_sentinel``.
na_sentinel : int, default -1
Value in ``codes`` to mark "not found".
Ignored when ``codes`` is None.
assume_unique : bool, default False
When True, ``values`` are assumed to be unique, which can speed up
the calculation. Ignored when ``codes`` is None.
verify : bool, default True
Check if codes are out of bound for the values and put out of bound
codes equal to na_sentinel. If ``verify=False``, it is assumed there
are no out of bound codes. Ignored when ``codes`` is None.
.. versionadded:: 0.25.0
Returns
-------
ordered : ndarray
Sorted ``values``
new_codes : ndarray
Reordered ``codes``; returned when ``codes`` is not None.
Raises
------
TypeError
* If ``values`` is not list-like or if ``codes`` is neither None
nor list-like
* If ``values`` cannot be sorted
ValueError
* If ``codes`` is not None and ``values`` contain duplicates.
"""
if not is_list_like(values):
raise TypeError(
"Only list-like objects are allowed to be passed to safe_sort as values"
)
if not isinstance(values, (np.ndarray, ABCExtensionArray)):
# don't convert to string types
dtype, _ = infer_dtype_from_array(values)
# error: Argument "dtype" to "asarray" has incompatible type "Union[dtype[Any],
# ExtensionDtype]"; expected "Union[dtype[Any], None, type, _SupportsDType, str,
# Union[Tuple[Any, int], Tuple[Any, Union[int, Sequence[int]]], List[Any],
# _DTypeDict, Tuple[Any, Any]]]"
values = np.asarray(values, dtype=dtype) # type: ignore[arg-type]
sorter = None
if (
not is_extension_array_dtype(values)
and lib.infer_dtype(values, skipna=False) == "mixed-integer"
):
ordered = _sort_mixed(values)
else:
try:
sorter = values.argsort()
ordered = values.take(sorter)
except TypeError:
# Previous sorters failed or were not applicable, try `_sort_mixed`
# which would work, but which fails for special case of 1d arrays
# with tuples.
if values.size and isinstance(values[0], tuple):
ordered = _sort_tuples(values)
else:
ordered = _sort_mixed(values)
# codes:
if codes is None:
return ordered
if not is_list_like(codes):
raise TypeError(
"Only list-like objects or None are allowed to "
"be passed to safe_sort as codes"
)
codes = ensure_platform_int(np.asarray(codes))
if not assume_unique and not len(unique(values)) == len(values):
raise ValueError("values should be unique if codes is not None")
if sorter is None:
# mixed types
hash_klass, values = get_data_algo(values)
t = hash_klass(len(values))
t.map_locations(values)
sorter = ensure_platform_int(t.lookup(ordered))
if na_sentinel == -1:
# take_nd is faster, but only works for na_sentinels of -1
order2 = sorter.argsort()
new_codes = take_nd(order2, codes, fill_value=-1)
if verify:
mask = (codes < -len(values)) | (codes >= len(values))
else:
mask = None
else:
reverse_indexer = np.empty(len(sorter), dtype=np.int_)
reverse_indexer.put(sorter, np.arange(len(sorter)))
# Out of bound indices will be masked with `na_sentinel` next, so we
# may deal with them here without performance loss using `mode='wrap'`
new_codes = reverse_indexer.take(codes, mode="wrap")
mask = codes == na_sentinel
if verify:
mask = mask | (codes < -len(values)) | (codes >= len(values))
if mask is not None:
np.putmask(new_codes, mask, na_sentinel)
return ordered, | ensure_platform_int(new_codes) | pandas.core.dtypes.common.ensure_platform_int |
import pandas as pd
import numpy as np
import datetime as dt
import pickle
import os
import shutil
import sys
from joblib import Parallel, delayed, cpu_count
import subprocess
from tqdm import tqdm
from copy import deepcopy
from blechpy.utils import print_tools as pt, write_tools as wt, userIO
from blechpy.utils.decorators import Logger
from blechpy.analysis import palatability_analysis as pal_analysis
from blechpy.analysis import spike_sorting as ss, spike_analysis, circus_interface as circ
from blechpy.analysis import blech_clustering as clust
from blechpy.plotting import palatability_plot as pal_plt, data_plot as datplt
from blechpy import dio
from blechpy.datastructures.objects import data_object
from blechpy.utils import spike_sorting_GUI as ssg
class dataset(data_object):
'''Stores information related to an intan recording directory, allows
executing basic processing and analysis scripts, and stores parameters data
for those analyses
Parameters
----------
file_dir : str (optional)
absolute path to a recording directory, if left empty a filechooser
will popup
'''
PROCESSING_STEPS = ['initialize parameters',
'extract_data', 'create_trial_list',
'mark_dead_channels',
'common_average_reference', 'spike_detection',
'spike_clustering', 'cleanup_clustering',
'sort_units', 'make_unit_plots',
'units_similarity', 'make_unit_arrays',
'make_psth_arrays', 'plot_psths',
'palatability_calculate', 'palatability_plot',
'overlay_psth']
def __init__(self, file_dir=None, data_name=None, shell=False):
'''Initialize dataset object from file_dir, grabs basename from name of
directory and initializes basic analysis parameters
Parameters
----------
file_dir : str (optional), file directory for intan recording data
Throws
------
ValueError
if file_dir is not provided and no directory is chosen
when prompted
NotADirectoryError : if file_dir does not exist
'''
super().__init__('dataset', file_dir, data_name=data_name, shell=shell)
h5_file = dio.h5io.get_h5_filename(self.root_dir)
if h5_file is None:
h5_file = os.path.join(self.root_dir, '%s.h5' % self.data_name)
print(f'No existing h5 file found. New h5 will be created at {h5_file}.')
else:
print(f'Existing h5 file found. Using {h5_file}.')
self.h5_file = h5_file
self.dataset_creation_date = dt.datetime.today()
# Outline standard processing pipeline and status check
self.processing_steps = dataset.PROCESSING_STEPS.copy()
self.process_status = dict.fromkeys(self.processing_steps, False)
def _change_root(self, new_root=None):
old_root = self.root_dir
new_root = super()._change_root(new_root)
self.h5_file = self.h5_file.replace(old_root, new_root)
return new_root
@Logger('Initializing Parameters')
def initParams(self, data_quality='clean', emg_port=None,
emg_channels=None, car_keyword=None,
car_group_areas=None,
shell=False, dig_in_names=None,
dig_out_names=None, accept_params=False):
'''
Initalizes basic default analysis parameters and allows customization
of parameters
Parameters (all optional)
-------------------------
data_quality : {'clean', 'noisy'}
keyword defining which default set of parameters to use to detect
headstage disconnection during clustering
default is 'clean'. Best practice is to run blech_clust as 'clean'
and re-run as 'noisy' if too many early cutoffs occurr
emg_port : str
Port ('A', 'B', 'C') of EMG, if there was an EMG. None (default)
will query user. False indicates no EMG port and not to query user
emg_channels : list of int
channel or channels of EMGs on port specified
default is None
car_keyword : str
Specifes default common average reference groups
defaults are found in CAR_defaults.json
Currently 'bilateral32' is only keyword available
If left as None (default) user will be queries to select common
average reference groups
shell : bool
False (default) for GUI. True for command-line interface
dig_in_names : list of str
Names of digital inputs. Must match number of digital inputs used
in recording.
None (default) queries user to name each dig_in
dig_out_names : list of str
Names of digital outputs. Must match number of digital outputs in
recording.
None (default) queries user to name each dig_out
accept_params : bool
True automatically accepts default parameters where possible,
decreasing user queries
False (default) will query user to confirm or edit parameters for
clustering, spike array and psth creation and palatability/identity
calculations
'''
# Get parameters from info.rhd
file_dir = self.root_dir
rec_info = dio.rawIO.read_rec_info(file_dir)
ports = rec_info.pop('ports')
channels = rec_info.pop('channels')
sampling_rate = rec_info['amplifier_sampling_rate']
self.rec_info = rec_info
self.sampling_rate = sampling_rate
# Get default parameters from files
clustering_params = dio.params.load_params('clustering_params', file_dir,
default_keyword=data_quality)
spike_array_params = dio.params.load_params('spike_array_params', file_dir)
psth_params = dio.params.load_params('psth_params', file_dir)
pal_id_params = dio.params.load_params('pal_id_params', file_dir)
spike_array_params['sampling_rate'] = sampling_rate
clustering_params['file_dir'] = file_dir
clustering_params['sampling_rate'] = sampling_rate
self.spike_array_params = spike_array_params
# Setup digital input mapping
if rec_info.get('dig_in'):
self._setup_digital_mapping('in', dig_in_names, shell)
dim = self.dig_in_mapping.copy()
else:
self.dig_in_mapping = None
if rec_info.get('dig_out'):
q = userIO.ask_user('Your info.rhd suggests you have digital '
'outputs. Is this True?', shell=shell)
if q == 1:
self._setup_digital_mapping('out', dig_out_names, shell)
dom = self.dig_out_mapping.copy()
else:
_ = rec_info.pop('dig_out')
self.dig_out_mapping = None
else:
self.dig_out_mapping = None
# Setup electrode and emg mapping
self._setup_channel_mapping(ports, channels, emg_port,
emg_channels, shell=shell)
# Set CAR groups
self._set_CAR_groups(group_keyword=car_keyword, group_areas=car_group_areas, shell=shell)
# Confirm parameters
if not accept_params:
conf = userIO.confirm_parameter_dict
clustering_params = conf(clustering_params,
'Clustering Parameters', shell=shell)
self.edit_spike_array_params(shell=shell)
psth_params = conf(psth_params,
'PSTH Parameters', shell=shell)
pal_id_params = conf(pal_id_params,
'Palatability/Identity Parameters\n'
'Valid unit_type is Single, Multi or All',
shell=shell)
# Store parameters
self.clustering_params = clustering_params
self.pal_id_params = pal_id_params
self.psth_params = psth_params
self._write_all_params_to_json()
self.process_status['initialize parameters'] = True
self.save()
def _set_CAR_groups(self, group_keyword=None, shell=False, group_areas=None):
'''Sets that electrode groups for common average referencing and
defines which brain region electrodes eneded up in
Parameters
----------
group_keyword : str or int
Keyword corresponding to a preset electrode grouping in CAR_params.json
Or integer indicating number of CAR groups
shell : bool
True for command-line interface, False (default) for GUI
'''
if not hasattr(self, 'electrode_mapping'):
raise ValueError('Set electrode mapping before setting CAR groups')
em = self.electrode_mapping.copy()
car_param_file = os.path.join(self.root_dir, 'analysis_params',
'CAR_params.json')
if os.path.isfile(car_param_file):
tmp = dio.params.load_params('CAR_params', self.root_dir)
if tmp is not None:
group_electrodes = tmp
else:
raise ValueError('CAR_params file exists in recording dir, but is empty')
else:
if group_keyword is None:
group_keyword = userIO.get_user_input(
'Input keyword for CAR parameters or number of CAR groups',
shell=shell)
if group_keyword is None:
ValueError('Must provide a keyword or number of groups')
if group_keyword.isnumeric():
num_groups = int(group_keyword)
group_electrodes = dio.params.select_CAR_groups(num_groups, em,
shell=shell)
else:
group_electrodes = dio.params.load_params('CAR_params',
self.root_dir,
default_keyword=group_keyword)
num_groups = len(group_electrodes)
if group_areas is not None and len(group_areas) == num_groups:
for i, x in enumerate(zip(group_electrodes, group_areas)):
em.loc[x[0], 'area'] = x[1]
em.loc[x[0], 'CAR_group'] = i
else:
group_names = ['Group %i' % i for i in range(num_groups)]
area_dict = dict.fromkeys(group_names, '')
area_dict = userIO.fill_dict(area_dict, 'Set Areas for CAR groups',
shell=shell)
for k, v in area_dict.items():
i = int(k.replace('Group', ''))
em.loc[group_electrodes[i], 'area'] = v
em.loc[group_electrodes[i], 'CAR_group'] = i
self.CAR_electrodes = group_electrodes
self.electrode_mapping = em.copy()
if os.path.isfile(self.h5_file):
dio.h5io.write_electrode_map_to_h5(self.h5_file, self.electrode_mapping)
@Logger('Re-labelling CAR group areas')
def set_electrode_areas(self, areas):
'''sets the electrode area for each CAR group.
Parameters
----------
areas : list of str
number of elements must match number of CAR groups
Throws
------
ValueError
'''
em = self.electrode_mapping.copy()
if len(em['CAR_group'].unique()) != len(areas):
raise ValueError('Number of items in areas must match number of CAR groups')
em['area'] = em['CAR_group'].apply(lambda x: areas[int(x)])
self.electrode_mapping = em.copy()
dio.h5io.write_electrode_map_to_h5(self.h5_file, self.electrode_mapping)
self.save()
def _setup_digital_mapping(self, dig_type, dig_in_names=None, shell=False):
'''sets up dig_in_mapping dataframe and queries user to fill in columns
Parameters
----------
dig_in_names : list of str (optional)
shell : bool (optional)
True for command-line interface
False (default) for GUI
'''
rec_info = self.rec_info
df = | pd.DataFrame() | pandas.DataFrame |
import os
import unittest
import numpy as np
import pandas as pd
from cgnal.core.data.model.ml import (
LazyDataset,
IterGenerator,
MultiFeatureSample,
Sample,
PandasDataset,
PandasTimeIndexedDataset,
CachedDataset,
features_and_labels_to_dataset,
)
from typing import Iterator, Generator
from cgnal.core.tests.core import TestCase, logTest
from tests import TMP_FOLDER
samples = [
Sample(features=[100, 101], label=1),
Sample(features=[102, 103], label=2),
Sample(features=[104, 105], label=3),
Sample(features=[106, 107], label=4),
Sample(features=[108, 109], label=5),
Sample(features=[110, 111], label=6),
Sample(features=[112, 113], label=7),
Sample(features=[114, 115], label=8),
Sample(features=[116, 117], label=9),
]
def samples_gen():
for sample in samples:
if not any([np.isnan(x).any() for x in sample.features]):
yield sample
lazyDat = LazyDataset(IterGenerator(samples_gen))
class features_and_labels_to_datasetTests(TestCase):
def test_features_and_labels_to_dataset(self):
dataset = features_and_labels_to_dataset(
pd.concat(
[
pd.Series([1, 0, 2, 3], name="feat1"),
| pd.Series([1, 2, 3, 4], name="feat2") | pandas.Series |
import pandas as pd
import datetime
from copy import deepcopy
from rgtfs import io, tables
def calculate_exits(row, calendar_dates_by_trip_id):
dow = {
0: "monday",
1: "tuesday",
2: "wednesday",
3: "thursday",
4: "friday",
5: "saturday",
6: "sunday",
}
_df = []
calendar = calendar_dates_by_trip_id.query(f'trip_id == "{row["trip_id"]}"').iloc[0]
current_date = pd.Timestamp(str(calendar["start_date"]))
end_date = pd.Timestamp(str(calendar["end_date"]))
# Loop through all dates in calendar
while current_date <= end_date:
# Has to be match day of the week
if not calendar[dow[current_date.weekday()]]:
current_date = current_date + datetime.timedelta(days=1)
current_time = pd.Timestamp(str(current_date.date()) + " " + row["start_time"])
end_time = pd.Timestamp(str(current_date.date()) + " " + row["end_time"])
while current_time < end_time:
_df.append(current_time)
current_time = current_time + datetime.timedelta(
seconds=row["headway_secs"]
)
current_date = current_date + datetime.timedelta(days=1)
_df = pd.DataFrame({"departure_datetime": _df})
_df["trip_id"] = row["trip_id"]
return _df
def generate_realized_trips_from_gtfs(gtfs_path):
"""Transforms a GTFS feed to realized_trips format (see README for specification).
It can either read a feed zip file or a folder.
Parameters
----------
gtfs_path : str
GTFS feed zip file or folder path
Returns
-------
pd.DataFrame
realized_trips data structure (see README for specification)
"""
gtfs = io.read_gtfs(gtfs_path, "km")
# Generates all exits
calendar_dates_by_trip_id = (
pd.merge(
gtfs.trips[["service_id", "trip_id"]], gtfs.calendar, on=["service_id"]
)
).drop_duplicates(subset=["service_id", "trip_id"])
realized_trips = []
for i, row in gtfs.frequencies.iterrows():
realized_trips.append(calculate_exits(row, calendar_dates_by_trip_id))
realized_trips = | pd.concat(realized_trips) | pandas.concat |
#!/usr/bin/env python3
import itertools
import string
from elasticsearch import Elasticsearch,helpers
import sys
import os
from glob import glob
import pandas as pd
import json
host = sys.argv[1]
port = int(sys.argv[2])
alias = sys.argv[3]
print(host)
print(port)
print(alias)
es = Elasticsearch([{'host': host, 'port': port}])
# create our test index
# Get all csv files in /root/data
files = [y for x in os.walk('/root/data') for y in glob(os.path.join(x[0], '*.csv'))]
count = 0
def clean_field(val):
val = val.split('.')
val = [i for i in val if i != '']
val = '_'.join(val)
val = val.split()
val = [i for i in val if i != '']
val = '_'.join(val)
val = val.split('/')
val = [i for i in val if i != '']
val = '_'.join(val)
return val
es.indices.delete(index=alias + '*', ignore=[400, 404])
indices = []
for file in files:
data = | pd.read_csv(file, sep=None, engine='python') | pandas.read_csv |
# -*- coding: utf-8 -*-
"""CARND3
Automatically generated by Colaboratory.
Original file is located at
https://colab.research.google.com/drive/1a44b45RBXDba9Nj99y_VLiS6lOwt5uKL
"""
import os
import csv
import cv2
import glob
from PIL import Image
import numpy as np
import sklearn
import random
import pandas as pd
from pandas import DataFrame
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
os.chdir(r"data")
# Import as a dataframe and plot steering
df = pd.read_csv('driving_log.csv', header=0)
df.columns = ["center_image", "left_image", "right_image", "steering", "throttle", "break", "speed"]
df.drop(['throttle', 'break', 'speed'], axis = 1, inplace = True)
import seaborn as sns
sns.set(style="whitegrid", color_codes=True)
sns.distplot(df['steering'], kde = False)
print(len(df))
## Oversample left and right turns. Downsample turns close to zero.
straight =[]
left_turn = []
right_turn = []
for i in range(len(df)):
keep_prob = random.random()
# Normal right turns - Double by adding small random fluctuations
if (df["steering"][i] >0.20 and df["steering"][i] <=0.50):
for j in range(2):
new_steering = df["steering"][i]*(1.0 + np.random.uniform(-1,1)/100.0)
right_turn.append([df["center_image"][i], df["left_image"][i], df["right_image"][i], new_steering])
# Normal left turns - Double by adding small random fluctuations
elif (df["steering"][i] >= -0.50 and df["steering"][i] < -0.15):
for j in range(2):
new_steering = df["steering"][i]*(1.0 + np.random.uniform(-1,1)/100.0)
left_turn.append([df["center_image"][i], df["left_image"][i], df["right_image"][i], new_steering])
## Zero angle steering - undersample by 10% worked best
elif (df["steering"][i] > -0.02 and df["steering"][i] < 0.02):
if keep_prob <=0.90:
straight.append([df["center_image"][i], df["left_image"][i], df["right_image"][i], df["steering"][i]])
else:
straight.append([df["center_image"][i], df["left_image"][i], df["right_image"][i], df["steering"][i]])
# Create a new list
new_list = []
new_list = right_turn + left_turn + straight
print(len(new_list), len(straight), len(left_turn), len(right_turn))
# Plot new distribution of steering
df_straight = pd.DataFrame(straight, columns=["center_image", "left_image", "right_image", "steering"])
df_left = pd.DataFrame(left_turn, columns=["center_image", "left_image", "right_image", "steering"])
df_right = | pd.DataFrame(right_turn, columns=["center_image", "left_image", "right_image", "steering"]) | pandas.DataFrame |
"""
Applying Box-Jenkins Forecasting Methodology
to Predict Massachusetts Cannabis Data
Copyright (c) 2021 Cannlytics and the Cannabis Data Science Meetup Group
Authors: <NAME> <<EMAIL>>
Created: 10/6/2021
Updated: 11/10/2021
License: MIT License <https://opensource.org/licenses/MIT>
References:
- Time Series forecasting using Auto ARIMA in Python
https://towardsdatascience.com/time-series-forecasting-using-auto-arima-in-python-bb83e49210cd
Data Sources:
MA Cannabis Control Commission
- Retail Sales by Date and Product Type: https://dev.socrata.com/foundry/opendata.mass-cannabis-control.com/xwf2-j7g9
- Approved Massachusetts Licensees: https://dev.socrata.com/foundry/opendata.mass-cannabis-control.com/hmwt-yiqy
- Average Monthly Price per Ounce for Adult-Use Cannabis: https://dev.socrata.com/foundry/opendata.mass-cannabis-control.com/rqtv-uenj
- Plant Activity and Volume: https://dev.socrata.com/foundry/opendata.mass-cannabis-control.com/j3q7-3usu
- Weekly sales by product type: https://dev.socrata.com/foundry/opendata.mass-cannabis-control.com/87rp-xn9v
Fed Fred
- MA Gross Domestic Product: https://fred.stlouisfed.org/series/MANQGSP
- MA Civilian Labor Force: https://fred.stlouisfed.org/series/MALF
- MA All Employees: https://fred.stlouisfed.org/series/MANA
- MA Avg. Weekly Wage: https://fred.stlouisfed.org/series/LES1252881600Q
- MA Minimum Wage: https://fred.stlouisfed.org/series/STTMINWGMA
- MA Population: https://fred.stlouisfed.org/series/MAPOP
"""
from dotenv import dotenv_values
import matplotlib.pyplot as plt
from matplotlib.ticker import FuncFormatter
import numpy as np
import pandas as pd
import pmdarima as pm
import requests
import seaborn as sns
import statsmodels.api as sm
# Internal imports
from utils import (
end_of_period_timeseries,
forecast_arima,
format_millions,
format_thousands,
reverse_dataframe,
set_training_period,
)
#--------------------------------------------------------------------------
# Get all MA public cannabis data.
#--------------------------------------------------------------------------
# Setup Socrata API, get the App Token, and define the headers.
config = dotenv_values('../.env')
app_token = config.get('APP_TOKEN', None)
headers = {'X-App-Token': app_token}
base = 'https://opendata.mass-cannabis-control.com/resource'
# Get production stats (total employees, total plants, etc.) j3q7-3usu
url = f'{base}/j3q7-3usu.json'
params = {'$limit': 2000, '$order': 'activitysummarydate DESC'}
response = requests.get(url, headers=headers, params=params)
production = pd.DataFrame(response.json(), dtype=float)
production = reverse_dataframe(production)
# Calculate sales difference.
production['sales'] = production['salestotal'].diff()
# FIX: Fix outlier that appears to have an extra 0.
outlier = production.loc[production.sales >= 10000000]
production.at[outlier.index, 'sales'] = 0
# FIX: Remove negative values.
negatives = production.loc[production.sales < 0]
production.at[negatives.index, 'sales'] = 0
# Aggregate daily production data into monthly and quarterly averages.
production['date'] = pd.to_datetime(production['activitysummarydate'])
production.set_index('date', inplace=True)
monthly_avg_production = production.resample('M').mean()
quarterly_avg_production = production.resample('Q').mean()
monthly_total_production = production.resample('M').sum()
quarterly_total_production = production.resample('Q').sum()
# Get licensees data.
url = f'{base}/hmwt-yiqy.json'
params = {'$limit': 10000, '$order': 'app_create_date DESC'}
response = requests.get(url, headers=headers, params=params)
licensees = pd.DataFrame(response.json(), dtype=float)
# Get the monthly average price per ounce.
url = f'{base}/rqtv-uenj.json'
params = {'$limit': 10000, '$order': 'date DESC'}
response = requests.get(url, headers=headers, params=params)
prices = pd.DataFrame(response.json(), dtype=float)
prices = reverse_dataframe(prices)
prices.set_index('date', inplace=True)
# Calculate the average price per specific quantity.
price_per_gram = prices.avg_1oz.astype(float).divide(28).round(2)
price_per_teenth = prices.avg_1oz.astype(float).divide(16).round(2)
price_per_eighth = prices.avg_1oz.astype(float).divide(8).round(2)
price_per_quarter = prices.avg_1oz.astype(float).divide(4).round(2)
# Get the products.
url = f'{base}/xwf2-j7g9.json'
params = {'$limit': 10000, '$order': 'saledate DESC'}
response = requests.get(url, headers=headers, params=params)
products = pd.DataFrame(response.json(), dtype=float)
products = reverse_dataframe(products)
products.set_index('saledate', inplace=True)
product_types = list(products.productcategoryname.unique())
#--------------------------------------------------------------------------
# Estimate historic sales per retialer, plants per cultivator,
# and employees per licensee.
#--------------------------------------------------------------------------
# Create weekly series.
weekly_sales = production.sales.resample('W-SUN').sum()
weekly_plants = production.total_planttrackedcount.resample('W-SUN').mean()
weekly_employees = production.total_employees.resample('W-SUN').mean()
# Find total retailers and cultivators.
retailers = licensees.loc[licensees.license_type == 'Marijuana Retailer']
cultivators = licensees.loc[licensees.license_type == 'Marijuana Cultivator']
total_retailers = len(retailers)
total_cultivators = len(cultivators)
total_licensees = len(licensees)
# Create total licensees series.
production['total_retailers'] = 0
production['total_cultivators'] = 0
production['total_licensees'] = 0
for index, _ in production.iterrows():
timestamp = index.isoformat()
production.at[index, 'total_retailers'] = len(licensees.loc[
(licensees.license_type == 'Marijuana Retailer') &
(licensees.app_create_date <= timestamp)
])
production.at[index, 'total_cultivators'] = len(licensees.loc[
(licensees.license_type == 'Marijuana Cultivator') &
(licensees.app_create_date <= timestamp)
])
production.at[index, 'total_licensees'] = len(licensees.loc[
(licensees.app_create_date <= timestamp)
])
# Create weekly averages.
weekly_total_retailers = production['total_retailers'].resample('W-SUN').mean()
weekly_total_cultivators = production['total_cultivators'].resample('W-SUN').mean()
weekly_total_licensees = production['total_licensees'].resample('W-SUN').mean()
# Estimate sales per retailer.
sales_per_retailer = weekly_sales / weekly_total_retailers
sales_per_retailer.plot()
plt.show()
# Estimate plants per cultivator.
plants_per_cultivator = weekly_plants / weekly_total_cultivators
plants_per_cultivator.plot()
plt.show()
# Estimate employees per licensee.
employees_per_license = weekly_employees / weekly_total_licensees
employees_per_license.plot()
plt.show()
#--------------------------------------------------------------------------
# Estimate sales, plants grown, and employees,
# total retailers, total cultivators, and total licensees
# in 2021 and 2022 using Box-Jenkins (ARIMA) methodology.
# Month fixed effects are used.
# Optional: Attempt to forecast with daily series with
# day-of-the-week fixed effects.
#--------------------------------------------------------------------------
# Specifiy training time periods.
train_start = '2020-06-01'
train_end = '2021-10-25'
# Define forecast horizon.
forecast_horizon = pd.date_range(
pd.to_datetime(train_end),
periods=62,
freq='w'
)
# Create month fixed effects (dummy variables),
# excluding 1 month (January) for comparison.
month_effects = pd.get_dummies(weekly_sales.index.month)
month_effects.index = weekly_sales.index
month_effects = set_training_period(month_effects, train_start, train_end)
forecast_month_effects = pd.get_dummies(forecast_horizon.month)
del month_effects[1]
try:
del forecast_month_effects[1]
except:
pass
# Forecast sales.
sales_model = pm.auto_arima(
set_training_period(weekly_sales, train_start, train_end),
X=month_effects,
start_p=0,
d=0,
start_q=0,
max_p=6,
max_d=6,
max_q=6,
seasonal=True,
start_P=0,
D=0,
start_Q=0,
max_P=6,
max_D=6,
max_Q=6,
information_criterion='bic',
alpha=0.2,
)
sales_forecast, sales_conf = forecast_arima(
sales_model,
forecast_horizon,
X=forecast_month_effects
)
# Forecast total plants.
plants_model = pm.auto_arima(
set_training_period(weekly_plants, train_start, train_end),
X=month_effects,
start_p=0,
d=0,
start_q=0,
max_p=6,
max_d=6,
max_q=6,
seasonal=True,
start_P=0,
D=0,
start_Q=0,
max_P=6,
max_D=6,
max_Q=6,
information_criterion='bic',
alpha=0.2,
)
plants_forecast, plants_conf = forecast_arima(
plants_model,
forecast_horizon,
X=forecast_month_effects
)
# Forecast total employees.
employees_model = pm.auto_arima(
set_training_period(weekly_employees, train_start, train_end),
X=month_effects,
start_p=0,
d=1,
start_q=0,
max_p=6,
max_d=6,
max_q=6,
seasonal=True,
start_P=0,
D=0,
start_Q=0,
max_P=6,
max_D=6,
max_Q=6,
information_criterion='bic',
alpha=0.2,
)
employees_forecast, employees_conf = forecast_arima(
employees_model,
forecast_horizon,
X=forecast_month_effects
)
# Forecast total retailers.
retailers_model = pm.auto_arima(
set_training_period(weekly_total_retailers, train_start, train_end),
X=month_effects,
start_p=0,
d=1,
start_q=0,
max_p=6,
max_d=6,
max_q=6,
seasonal=True,
start_P=0,
D=0,
start_Q=0,
max_P=6,
max_D=6,
max_Q=6,
information_criterion='bic',
alpha=0.2,
# m=12,
)
total_retailers_forecast, total_retailers_conf = forecast_arima(
retailers_model,
forecast_horizon,
X=forecast_month_effects
)
# Forecast total cultivators.
cultivators_model = pm.auto_arima(
set_training_period(weekly_total_cultivators, train_start, train_end),
X=month_effects,
start_p=0,
d=1,
start_q=0,
max_p=6,
max_d=6,
max_q=6,
seasonal=True,
start_P=0,
D=0,
start_Q=0,
max_P=6,
max_D=6,
max_Q=6,
information_criterion='bic',
alpha=0.2,
# m=12,
)
total_cultivators_forecast, total_cultivators_conf = forecast_arima(
cultivators_model,
forecast_horizon,
X=forecast_month_effects
)
# Forecast total licensees.
licensees_model = pm.auto_arima(
set_training_period(weekly_total_licensees, train_start, train_end),
X=month_effects,
start_p=0,
d=1,
start_q=0,
max_p=6,
max_d=6,
max_q=6,
seasonal=True,
start_P=0,
D=0,
start_Q=0,
max_P=6,
max_D=6,
max_Q=6,
information_criterion='bic',
alpha=0.2,
# m=12,
)
total_licensees_forecast, total_licensees_conf = forecast_arima(
licensees_model,
forecast_horizon,
X=forecast_month_effects
)
# Predict total sales per retailer in 2022.
# TODO: Figure out how to estimate confidence bounds?
forecast_sales_per_retailer = sales_forecast / total_retailers_forecast
# Predict total plants per cultivator in 2022.
forecast_plants_per_cultivator = plants_forecast / total_cultivators_forecast
# Predict total employees per licensee in 2022.
forecast_employees_per_license = employees_forecast / total_licensees_forecast
#--------------------------------------------------------------------------
# Visualize the forecasts as 2x3 time series plots.
# Sales | Plants
# Retailers | Cultivators
# Sales per Retailer | Plants per Cultivator
#--------------------------------------------------------------------------
def plot_forecast(
ax,
forecast,
historic=None,
conf=None,
title=None,
color=None,
formatter=None,
):
"""Plot a time series forecast.
Args:
ax (): The axes on which to plot the forecast.
forecast (Series): The forecast to plot.
historic (Series): Optional historic time series to plot.
conf (Array): An optional 2xN array of lower and upper confidence
bounds for the forecast series.
title (str): An optional title to place above the chart.
color: (str): An optional color hex code.
formatter (func): An optional formatting function for the Y axis.
"""
forecast.plot(color=color, style='--', label='Forecast')
if conf is not None:
plt.fill_between(
forecast.index,
conf[:, 0],
conf[:, 1],
alpha=0.1,
color=color,
)
if historic is not None:
historic.plot(color=color, label='Historic')
if title is not None:
plt.title(title, fontsize=24, pad=10)
if formatter is not None:
yaxis_format = FuncFormatter(formatter)
ax.yaxis.set_major_formatter(yaxis_format)
plt.gca().set(ylim=0)
plt.setp(ax.get_yticklabels()[0], visible=False)
plt.xlabel('')
plt.xticks(fontsize=18)
plt.yticks(fontsize=18)
# Define the plot style.
plt.style.use('fivethirtyeight')
plt.rcParams['font.family'] = 'Times New Roman'
palette = sns.color_palette('tab10')
# Plot all series.
fig = plt.figure(figsize=(40, 25))
# Plot sales.
ax1 = plt.subplot(3, 2, 1)
plot_forecast(
ax1,
sales_forecast,
historic=weekly_sales,
conf=sales_conf,
title='Cannabis Sales',
color=palette[0],
formatter=format_millions,
)
# Plot plants.
ax2 = plt.subplot(3, 2, 2)
plot_forecast(
ax2,
plants_forecast,
historic=weekly_plants,
conf=plants_conf,
title='Cannabis Plants',
color=palette[-1],
formatter=format_thousands,
)
# Plot retailers.
ax3= plt.subplot(3, 2, 3)
plot_forecast(
ax3,
total_retailers_forecast,
historic=weekly_total_retailers,
conf=total_retailers_conf,
title='Cannabis Retailers',
color=palette[3],
# formatter=format_thousands,
)
# Plot cultivators.
ax4= plt.subplot(3, 2, 4)
plot_forecast(
ax4,
total_cultivators_forecast,
historic=weekly_total_cultivators,
conf=total_cultivators_conf,
title='Cannabis Cultivators',
color=palette[2],
# formatter=format_thousands,
)
# Plot average sales per retailer.
ax5= plt.subplot(3, 2, 5)
plot_forecast(
ax5,
forecast_sales_per_retailer,
historic=sales_per_retailer,
# conf=total_cultivators_conf,
title='Average Cannabis Sales per Retailer',
color=palette[4],
formatter=format_thousands,
)
# Plot average plants per cultivator.
ax6 = plt.subplot(3, 2, 6)
plot_forecast(
ax6,
forecast_plants_per_cultivator,
historic=plants_per_cultivator,
# conf=total_cultivators_conf,
title='Average Cannabis Plants per Cultivator',
color=palette[5],
# formatter=format_thousands,
)
# Add figure notes.
notes = """Data: Cannabis sales, total tracked plants, and licensees data from October 15, 2018 through October 26, 2021.
Data Source: MA Cannabis Control Commission."""
plt.figtext(0.05, -0.01, notes, ha='left', fontsize=16)
# Add a title above all of the subplots.
fig.suptitle(
'Massachusetts Historic and Predicted Cannabis Market Performance',
fontsize=40
)
# Save the figure.
plt.margins(1, 1)
plt.savefig(
'figures/ma_market_forecast.pdf',
dpi=300,
bbox_inches='tight',
pad_inches=0.75,
transparent=True,
)
# Show the figure.
plt.show()
# Optional: Show a legend?
# plt.legend(loc='lower left', fontsize=18)
# Optional: Find a clever way to display 2022 forecast totals (curly braces?)
#--------------------------------------------------------------------------
# Optional: Estimate a production function with the forecasted values
# and calculate the estimated competitive wage and interest rate,
# getting supplemental data from FRED (Federal Reserve Economic Data).
#--------------------------------------------------------------------------
# Initialize Fred client.
from fredapi import Fred
config = dotenv_values('../.env')
fred = Fred(api_key=config.get('FRED_API_KEY'))
# Find the observation time start.
observation_start = production.index.min().isoformat()
# Get average weekly hours worked in MA.
avg_weekly_hours = fred.get_series('SMU25000000500000002SA', observation_start=observation_start)
avg_weekly_hours = end_of_period_timeseries(avg_weekly_hours)
avg_weekly_hours = avg_weekly_hours.resample('W-Sun').ffill().iloc[:-1]
#--------------------------------------------------------------------------
# Optional: Estimate historic competitive wages and interest rates.
#--------------------------------------------------------------------------
# Define variables.
Y = weekly_sales
K = weekly_plants
L = weekly_employees * avg_weekly_hours
# Exclude missing observations.
missing_sales = Y.loc[Y == 0].index
Y = Y[~Y.index.isin(missing_sales)]
K = K[~K.index.isin(missing_sales)]
L = L[~L.index.isin(missing_sales)]
# Restrict time frame.
Y = Y.loc[(Y.index >= pd.to_datetime('2019-01-01')) & (Y.index <= pd.to_datetime('2021-08-01'))]
K = K.loc[(K.index >= pd.to_datetime('2019-01-01')) & (K.index <= pd.to_datetime('2021-08-01'))]
L = L.loc[(L.index >= | pd.to_datetime('2019-01-01') | pandas.to_datetime |
"""Unit tests for engine module utility functions."""
import numpy as np
import pandas as pd
import pytest
from pandera.engines import utils
@pytest.mark.parametrize(
"data_container, data_type, expected_failure_cases",
[
[pd.Series(list("ab1cd3")), int, [False, False, True] * 2],
[pd.Series(list("12345")), int, [True] * 5],
[ | pd.Series([1, 2, "foo", "bar"]) | pandas.Series |
# coding=utf-8
import pandas as pd
import xgboost as xgb
from sklearn.metrics import f1_score
import param
############################ 定义评估函数 ############################
def micro_avg_f1(preds, dtrain):
y_true = dtrain.get_label()
return 'micro_avg_f1', f1_score(y_true, preds, average='micro')
############################ 加载特征 & 标签 ############################
df_tfidf_lr = pd.read_csv(param.data_path + '/output/feature/tfidf/lr_prob_12w.csv')
df_tfidf_bnb = pd.read_csv(param.data_path + '/output/feature/tfidf/bnb_prob_12w.csv')
df_tfidf_mnb = | pd.read_csv(param.data_path + '/output/feature/tfidf/mnb_prob_12w.csv') | pandas.read_csv |
from datetime import datetime, timedelta
from importlib import reload
import string
import sys
import numpy as np
import pytest
from pandas._libs.tslibs import iNaT
from pandas.core.dtypes.dtypes import CategoricalDtype
import pandas as pd
from pandas import (
Categorical,
DataFrame,
Index,
Series,
Timedelta,
Timestamp,
date_range,
)
import pandas._testing as tm
class TestSeriesDtypes:
def test_dt64_series_astype_object(self):
dt64ser = Series( | date_range("20130101", periods=3) | pandas.date_range |
import os
from pathlib import Path
import joblib
import pandas as pd
import numpy as np
from multiprocessing import Pool
from collections import defaultdict
import functools
import re
import sys
sys.path.insert(0, './code')
from utils import DataLogger # noqa: E402
class DataNotFoundException(Exception):
pass
def get_time_split(df):
df_12 = df[df['dt'] <= 12]
df_16 = df[(df['dt'] > 12) & (df['dt'] <= 16)]
# df_20 = df[(df['dt'] > 16) & (df['dt'] <= 19)]
# df_21 = df[(df['dt'] > 17) & (df['dt'] <= 20)]
df_22 = df[(df['dt'] > 18) & (df['dt'] <= 21)]
df_23 = df[(df['dt'] > 19) & (df['dt'] <= 22)]
# df_24 = df[(df['dt'] > 20) & (df['dt'] <= 23)]
# df_25 = df[(df['dt'] > 21) & (df['dt'] <= 24)]
r_dict = {
"one_to_twelve": df_12,
"twelve_to_sixteen": df_16,
# "prev_three_months_20": df_20,
# "prev_three_months_21": df_21,
"prev_three_months_22": df_22,
"prev_three_months_23": df_23,
# "prev_three_months_24": df_24,
# "prev_three_months_25": df_25
}
return r_dict
def get_merge_dict():
merge_dict = {
# 20: ["one_to_twelve", "twelve_to_sixteen", "prev_three_months_20"],
# 21: ["one_to_twelve", "twelve_to_sixteen", "prev_three_months_21"],
22: ["one_to_twelve", "twelve_to_sixteen", "prev_three_months_22"],
23: ["one_to_twelve", "twelve_to_sixteen", "prev_three_months_23"],
# 24: ["one_to_twelve", "twelve_to_sixteen", "prev_three_months_24"],
# 25: ["one_to_twelve", "twelve_to_sixteen", "prev_three_months_25"],
}
return merge_dict
def get_time_split_result(a_func):
@functools.wraps(a_func)
def wrapper(self, df):
r_dict = defaultdict(list)
df_dict = get_time_split(df)
use_dict = {key: a_func(self, df_dict[key]) for key in df_dict.keys()}
merge_dict = get_merge_dict()
for dt in merge_dict.keys():
vals_12 = use_dict[merge_dict[dt][0]]
vals_16 = use_dict[merge_dict[dt][1]]
vals_prevs = use_dict[merge_dict[dt][2]]
for val, val_12, val_16 in zip(vals_prevs, vals_12, vals_16):
name = val[0]
name_12 = "{}_12".format(name)
name_16 = "{}_16".format(name)
r_dict[name].append(val[1])
r_dict[name_12].append(val_12[1])
r_dict[name_16].append(val_16[1])
return r_dict
return wrapper
class DataLoader():
def __init__(self):
self.output_path = Path(os.path.abspath(os.getcwd())) / 'output'
self.input_path = Path(os.path.abspath(os.getcwd())) / 'input'
self.model_path = Path(os.path.abspath(os.getcwd())) / 'model'
def save_data(self, cls, data_name, message):
logger = DataLogger()
logger.save_data("Save data {} is generated from {}".format(
data_name, message))
joblib.dump(cls, self.output_path / data_name)
logger.save_data("{} is sucessfully saved".format(data_name))
def load_data(self, data_name, data_type='joblib', **kwargs):
if data_type == 'joblib':
data = joblib.load(self.input_path / data_name, **kwargs)
elif data_type == 'csv':
data = pd.read_csv(self.input_path / data_name, **kwargs)
return data
def load_result(self, data_name, data_type='joblib', **kwargs):
if data_type == 'joblib':
data = joblib.load(self.output_path / data_name, **kwargs)
elif data_type == 'csv':
data = | pd.read_csv(self.output_path / data_name, **kwargs) | pandas.read_csv |
import time
import pandas as pd
import A01_process_route_seq
import A02_process_package_data
import A03_process_route_data
import numpy as np
import os
import json
def haversine_np(lon1, lat1, lon2, lat2):
"""
Calculate the great circle distance between two points
on the earth (specified in decimal degrees)
All args must be of equal length.
"""
lon1, lat1, lon2, lat2 = map(np.radians, [lon1, lat1, lon2, lat2])
dlon = lon2 - lon1
dlat = lat2 - lat1
a = np.sin(dlat/2.0)**2 + np.cos(lat1) * np.cos(lat2) * np.sin(dlon/2.0)**2
c = 2 * np.arcsin(np.sqrt(a))
km = 6367 * c
return km
def merge_all_data(route_data, seq_df, package, save_file, data_output_path, data_apply_output_path, data_path):
#tic = time.time()
#print('load data time',time.time() - tic)
assert len(route_data.loc[route_data['stops'].isna()]) == 0
assert len(seq_df.loc[seq_df['stops'].isna()]) == 0
assert len(package.loc[package['stops'].isna()]) == 0
#%%
route_seq = pd.merge(route_data, seq_df,on = ['route_id','stops'], how = 'left')
route_seq = route_seq.sort_values(['route_id','seq_ID'])
assert len(route_seq.loc[route_seq['seq_ID'].isna()]) == 0
all_zone_id = list(pd.unique(route_seq['zone_id']))
all_zone_id = [x for x in all_zone_id if str(x) != 'nan']
if 'INIT' in all_zone_id:
all_zone_id.remove('INIT')
replace_zone = []
# key = all_zone_id[0]
for key in all_zone_id:
try:
split_dot_1, split_dot_2 = key.split('.')
split_dot_1_1, split_dot_1_2 = split_dot_1.split('-')
assert len(split_dot_1_1) == 1
assert len(split_dot_1_2) > 0
assert len(split_dot_2) == 2
except:
replace_zone.append(key)
# check
###
#z = replace_zone[1]
all_routes_with_bad_zone = list(pd.unique(route_seq.loc[route_seq['zone_id'].isin(replace_zone),'route_id']))
for r in all_routes_with_bad_zone:
info = route_seq.loc[route_seq['route_id'] == r].copy()
info_with_invalid_zone = info.loc[info['zone_id'].isin(replace_zone)]
prop_inv_zone = len(info_with_invalid_zone)/len(info)
if prop_inv_zone > 0.3:
#in_valid_zone = list(pd.unique([''])) # not fill
continue
else:
route_seq.loc[(route_seq['route_id'] == r) & (route_seq['zone_id'].isin(replace_zone)),'zone_id'] = np.nan
#,'zone_id'] = np.nan
###
#a1=1
#print(route_seq['route_id'].iloc[0])
# route_seq.loc[route_seq['route_id'] == 'RouteID_00143bdd-0a6b-49ec-bb35-36593d303e77', 'zone_id'] = np.nan
route_seq.loc[route_seq['type'] == 'Station', 'zone_id'] = 'INIT'
# first fill all na routes
route_seq['zone_na'] = route_seq['zone_id'].isna()
route_seq['max_stops_num_except_ini'] = route_seq.groupby(['route_id'])['stops'].transform('count') - 1
route_seq['zone_na_num_stops'] = route_seq.groupby(['route_id'])['zone_na'].transform('sum')
route_all_na = route_seq.loc[route_seq['max_stops_num_except_ini']==route_seq['zone_na_num_stops']]
route_all_na_id = list(pd.unique(route_all_na['route_id']))
for key in route_all_na_id:
route_seq.loc[(route_seq['route_id'] == key)&(route_seq['zone_id'] != 'INIT'), 'zone_id'] = 'A-999.9Z'
route_seq = route_seq.drop(columns=['zone_na','max_stops_num_except_ini','zone_na_num_stops'])
##############
FILL_NA_METHOD = 'Nearest_TT_N' # NEAREST:
if FILL_NA_METHOD == 'Nearest':
##########fill by nearest
na_stop = route_seq.loc[route_seq['zone_id'].isna(),['route_id','stops','lat','lng']]
no_na_stop = route_seq.loc[(~(route_seq['zone_id'].isna())) & (route_seq['zone_id']!='INIT'),['route_id','stops','lat','lng','zone_id']]
no_na_stop = no_na_stop.rename(columns = {'stops':'nearby_stops','lat':'lat_nearby','lng':'lng_nearby'})
na_stop_nearby_stop = na_stop.merge(no_na_stop, on = ['route_id'])
na_stop_nearby_stop['dist'] = haversine_np(na_stop_nearby_stop['lng'].values, na_stop_nearby_stop['lat'].values,
na_stop_nearby_stop['lng_nearby'].values, na_stop_nearby_stop['lat_nearby'].values)
na_stop_nearby_stop = na_stop_nearby_stop.sort_values(['route_id','stops','dist'])
na_stop_nearby_stop_with_zone = na_stop_nearby_stop.groupby(['route_id','stops'], as_index = False).first()
na_stop_nearby_stop_with_zone['zone_id_infer'] = na_stop_nearby_stop_with_zone['zone_id']
route_seq = route_seq.merge(na_stop_nearby_stop_with_zone[['route_id','stops','zone_id_infer']], on = ['route_id','stops'], how = 'left')
route_seq.loc[route_seq['zone_id'].isna(),'zone_id'] = route_seq.loc[route_seq['zone_id'].isna(),'zone_id_infer']
route_seq = route_seq.drop(columns = ['zone_id_infer'])
#check_na = route_seq.loc[route_seq['zone_id'].isna()]
#a=1
#check_all_na = na_stop_nearby_stop.loc[na_stop_nearby_stop['']]
a=1
route_seq_fill_na = route_seq.copy()
elif FILL_NA_METHOD == 'Nearest_N':
near_num = 2
##########fill by nearest
na_stop = route_seq.loc[route_seq['zone_id'].isna(),['route_id','stops','lat','lng']]
no_na_stop = route_seq.loc[(~(route_seq['zone_id'].isna())) & (route_seq['zone_id']!='INIT'),['route_id','stops','lat','lng','zone_id']]
no_na_stop = no_na_stop.rename(columns = {'stops':'nearby_stops','lat':'lat_nearby','lng':'lng_nearby'})
na_stop_nearby_stop = na_stop.merge(no_na_stop, on = ['route_id'])
na_stop_nearby_stop['dist'] = haversine_np(na_stop_nearby_stop['lng'].values, na_stop_nearby_stop['lat'].values,
na_stop_nearby_stop['lng_nearby'].values, na_stop_nearby_stop['lat_nearby'].values)
na_stop_nearby_stop = na_stop_nearby_stop.sort_values(['route_id','stops','dist'])
na_stop_nearby_stop_with_zone = na_stop_nearby_stop.groupby(['route_id','stops'], as_index = False).head(near_num)
na_stop_nearby_stop_with_zone = na_stop_nearby_stop_with_zone.reset_index(drop=True)
na_stop_nearby_stop_with_zone['weight'] = 1/ na_stop_nearby_stop_with_zone['dist']
na_stop_nearby_stop_with_zone_num = na_stop_nearby_stop_with_zone.groupby(['route_id','stops','zone_id'])['weight'].sum().reset_index()
na_stop_nearby_stop_with_zone_num = na_stop_nearby_stop_with_zone_num.rename(columns = {'weight':'num_fre_zone'})
na_stop_nearby_stop_with_zone_num = na_stop_nearby_stop_with_zone_num.sort_values(['route_id','stops','num_fre_zone'],ascending=False)
na_stop_nearby_stop_with_zone = na_stop_nearby_stop_with_zone_num.groupby(['route_id','stops']).first().reset_index()
na_stop_nearby_stop_with_zone['zone_id_infer'] = na_stop_nearby_stop_with_zone['zone_id']
route_seq = route_seq.merge(na_stop_nearby_stop_with_zone[['route_id','stops','zone_id_infer']], on = ['route_id','stops'], how = 'left')
route_seq.loc[route_seq['zone_id'].isna(),'zone_id'] = route_seq.loc[route_seq['zone_id'].isna(),'zone_id_infer']
route_seq = route_seq.drop(columns = ['zone_id_infer'])
#check_na = route_seq.loc[route_seq['zone_id'].isna()]
#a=1
#check_all_na = na_stop_nearby_stop.loc[na_stop_nearby_stop['']]
a=1
route_seq_fill_na = route_seq.copy()
elif FILL_NA_METHOD == 'Nearest_TT_N':
near_num = 1
filepath = data_path + 'model_apply_inputs/new_travel_times.json'
with open(filepath, newline='') as in_file:
travel_time_matrix = json.load(in_file)
stop_tt = {'route_id': [], 'from_stop': [], 'to_stop': [], 'travel_time': []}
for key in travel_time_matrix:
for from_zone in travel_time_matrix[key]:
for to_zone in travel_time_matrix[key][from_zone]:
if (from_zone != to_zone): # and (from_zone == 'INIT'):
stop_tt['route_id'].append(key)
stop_tt['from_stop'].append(from_zone)
stop_tt['to_stop'].append(to_zone)
stop_tt['travel_time'].append(travel_time_matrix[key][from_zone][to_zone])
stop_tt_df = | pd.DataFrame(stop_tt) | pandas.DataFrame |
#import urllib2
import csv
import sys
import re
from datetime import datetime
import time
import pandas as pd
import configparser
import hashlib
import os
import rdflib
import logging
logging.getLogger().disabled = True
if sys.version_info[0] == 3:
from importlib import reload
reload(sys)
if sys.version_info[0] == 2:
sys.setdefaultencoding('utf8')
whyis = rdflib.Namespace('http://vocab.rpi.edu/whyis/')
np = rdflib.Namespace("http://www.nanopub.org/nschema#")
prov = rdflib.Namespace("http://www.w3.org/ns/prov#")
dc = rdflib.Namespace("http://purl.org/dc/terms/")
sio = rdflib.Namespace("http://semanticscience.org/resource/")
setl = rdflib.Namespace("http://purl.org/twc/vocab/setl/")
pv = rdflib.Namespace("http://purl.org/net/provenance/ns#")
skos = rdflib.Namespace("http://www.w3.org/2008/05/skos#")
rdfs = rdflib.RDFS
rdf = rdflib.RDF
owl = rdflib.OWL
xsd = rdflib.XSD
def parseString(input_string, delim) :
my_list = input_string.split(delim)
for i in range(0,len(my_list)) :
my_list[i] = my_list[i].strip()
return my_list
def codeMapper(input_word) :
unitVal = input_word
for unit_label in unit_label_list :
if (unit_label == input_word) :
unit_index = unit_label_list.index(unit_label)
unitVal = unit_uri_list[unit_index]
for unit_code in unit_code_list :
if (unit_code == input_word) :
unit_index = unit_code_list.index(unit_code)
unitVal = unit_uri_list[unit_index]
return unitVal
def convertImplicitToKGEntry(*args) :
if (args[0][:2] == "??") :
if (studyRef is not None ) :
if (args[0]==studyRef) :
return "<" + prefixes[kb] + args[0][2:] + ">"
if (len(args) == 2) :
return "<" + prefixes[kb] + args[0][2:] + "-" + args[1] + ">"
else :
return "<" + prefixes[kb] + args[0][2:] + ">"
elif (':' not in args[0]) :
# Check for entry in column list
for item in explicit_entry_list :
if args[0] == item.Column :
if (len(args) == 2) :
return "<" + prefixes[kb] + args[0].replace(" ","_").replace(",","").replace("(","").replace(")","").replace("/","-").replace("\\","-") + "-" + args[1] + ">"
else :
return "<" + prefixes[kb] + args[0].replace(" ","_").replace(",","").replace("(","").replace(")","").replace("/","-").replace("\\","-") + ">"
return '"' + args[0] + "\"^^xsd:string"
else :
return args[0]
def checkImplicit(input_word) :
try:
if (input_word[:2] == "??") :
return True
else :
return False
except Exception as e:
print("Something went wrong in checkImplicit()" + str(e))
sys.exit(1)
def isfloat(term):
try:
float(term)
return True
except ValueError:
return False
def isURI(term):
try:
if any(c in term for c in ("http://","https://")) :
return True
else:
return False
except ValueError:
return False
def isSchemaVar(term) :
for entry in explicit_entry_list :
if term == entry[1] :
return True
return False
def assignVID(implicit_entry_tuples,timeline_tuple,a_tuple,column, npubIdentifier) :
v_id = npubIdentifier
for v_tuple in implicit_entry_tuples : # referenced in implicit list
if v_tuple["Column"] == a_tuple[column]:
v_id = hashlib.md5((str(v_tuple) + str(npubIdentifier)).encode("utf-8")).hexdigest()
if v_id == None : # maybe it's referenced in the timeline
for t_tuple in timeline_tuple:
if t_tuple["Column"] == a_tuple[column]:
#print("Got here")
v_id = hashlib.md5((str(t_tuple) + str(npubIdentifier)).encode("utf-8")).hexdigest()
if v_id == npubIdentifier : # if it's not in implicit list or timeline
print("Warning, " + column + " ID assigned to nanopub ID")
return v_id
def assignTerm(col_headers, column, implicit_entry_tuples, a_tuple, row, v_id) :
termURI = None
for v_tuple in implicit_entry_tuples : # referenced in implicit list
if v_tuple["Column"] == a_tuple[column]:
if "Template" in v_tuple :
template_term = extractTemplate(col_headers,row,v_tuple["Template"])
termURI = "<" + prefixes[kb] + template_term + ">"
if termURI is None :
termURI = convertImplicitToKGEntry(a_tuple[column],v_id)
return termURI
'''def processPrefixes(output_file,query_file):
if 'prefixes' in config['Prefixes']:
prefix_fn = config['Prefixes']['prefixes']
else:
prefix_fn="prefixes.txt"
prefix_file = open(prefix_fn,"r")
prefixes = prefix_file.readlines()
for prefix in prefixes :
#print(prefix.find(">"))
output_file.write(prefix)
query_file.write(prefix[1:prefix.find(">")+1])
query_file.write("\n")
prefix_file.close()
output_file.write("\n")'''
def checkTemplate(term) :
if "{" in term and "}" in term:
return True
return False
def extractTemplate(col_headers,row,term) :
while checkTemplate(term) :
open_index = term.find("{")
close_index = term.find("}")
key = term[open_index+1:close_index]
term = term[:open_index] + str(row[col_headers.index(key)+1]) + term[close_index+1:]
return term
def extractExplicitTerm(col_headers,row,term) : # need to write this function
while checkTemplate(term) :
open_index = term.find("{")
close_index = term.find("}")
key = term[open_index+1:close_index]
if isSchemaVar(key) :
for entry in explicit_entry_list :
if entry.Column == key :
if pd.notnull(entry.Template) :
term = extractTemplate(col_headers,row,entry.Template)
else :
typeString = ""
if pd.notnull(entry.Attribute) :
typeString += str(entry.Attribute)
if pd.notnull(entry.Entity) :
typeString += str(entry.Entity)
if pd.notnull(entry.Label) :
typeString += str(entry.Label)
if pd.notnull(entry.Unit) :
typeString += str(entry.Unit)
if pd.notnull(entry.Time) :
typeString += str(entry.Time)
if pd.notnull(entry.inRelationTo) :
typeString += str(entry.inRelationTo)
if pd.notnull(entry.wasGeneratedBy) :
typeString += str(entry.wasGeneratedBy)
if pd.notnull(entry.wasDerivedFrom) :
typeString += str(entry.wasDerivedFrom)
identifierKey = hashlib.md5((str(row[col_headers.index(key)+1])+typeString).encode("utf-8")).hexdigest()
term = entry.Column + "-" + identifierKey
#return extractTemplate(col_headers,row,entry.Template)
else : # What does it mean for a template reference to not be a schema variable?
print("Warning: Template reference " + term + " is not be a schema variable")
term = term[:open_index] + str(row[col_headers.index(key)+1]) + term[close_index+1:] # Needs updating probably, at least checking
return term
def writeClassAttributeOrEntity(item, term, input_tuple, assertionString, whereString, swrlString) :
if (pd.notnull(item.Entity)) and (pd.isnull(item.Attribute)) :
if ',' in item.Entity :
entities = parseString(item.Entity,',')
for entity in entities :
assertionString += " ;\n <" + rdfs.subClassOf + "> " + codeMapper(entity)
whereString += codeMapper(entity) + " "
swrlString += codeMapper(entity) + "(" + term + ") ^ "
if entities.index(entity) + 1 != len(entities) :
whereString += ", "
else :
assertionString += " ;\n <" + rdfs.subClassOf + "> " + codeMapper(item.Entity)
whereString += codeMapper(item.Entity) + " "
swrlString += codeMapper(item.Entity) + "(" + term + ") ^ "
input_tuple["Entity"]=codeMapper(item.Entity)
if (input_tuple["Entity"] == "hasco:Study") :
global studyRef
studyRef = item.Column
input_tuple["Study"] = item.Column
elif (pd.isnull(item.Entity)) and (pd.notnull(item.Attribute)) :
if ',' in item.Attribute :
attributes = parseString(item.Attribute,',')
for attribute in attributes :
assertionString += " ;\n <" + rdfs.subClassOf + "> " + codeMapper(attribute)
whereString += codeMapper(attribute) + " "
swrlString += codeMapper(attribute) + "(" + term + ") ^ "
if attributes.index(attribute) + 1 != len(attributes) :
whereString += ", "
else :
assertionString += " ;\n <" + rdfs.subClassOf + "> " + codeMapper(item.Attribute)
whereString += codeMapper(item.Attribute) + " "
swrlString += codeMapper(item.Attribute) + "(" + term + ") ^ "
input_tuple["Attribute"]=codeMapper(item.Attribute)
else :
print("Warning: Entry not assigned an Entity or Attribute value, or was assigned both.")
input_tuple["Attribute"]=codeMapper("sio:Attribute")
assertionString += " ;\n <" + rdfs.subClassOf + "> sio:Attribute"
whereString += "sio:Attribute "
swrlString += "sio:Attribute(" + term + ") ^ "
return [input_tuple, assertionString, whereString, swrlString]
def writeClassAttributeOf(item, term, input_tuple, assertionString, whereString, swrlString) :
if (pd.notnull(item.attributeOf)) :
if checkTemplate(item.attributeOf) :
open_index = item.attributeOf.find("{")
close_index = item.attributeOf.find("}")
key = item.attributeOf[open_index+1:close_index]
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + "> " + convertImplicitToKGEntry(key) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["attributeOf"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["attributeOf"] + "> " + convertImplicitToKGEntry(key)
whereString += " ;\n <" + properties_tuple["attributeOf"] + "> ?" + key.lower() + "_E"
swrlString += properties_tuple["attributeOf"] + "(" + term + " , " + [key,key[1:] + "_V"][checkImplicit(key)] + ") ^ "
else :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + "> " + convertImplicitToKGEntry(item.attributeOf) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["attributeOf"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["attributeOf"] + "> " + convertImplicitToKGEntry(item.attributeOf)
whereString += " ;\n <" + properties_tuple["attributeOf"] + "> " + [item.attributeOf + " ",item.attributeOf[1:] + "_V "][checkImplicit(item.attributeOf)]
swrlString += properties_tuple["attributeOf"] + "(" + term + " , " + [item.attributeOf,item.attributeOf[1:] + "_V"][checkImplicit(item.attributeOf)] + ") ^ "
input_tuple["isAttributeOf"]=item.attributeOf
return [input_tuple, assertionString, whereString, swrlString]
def writeClassUnit(item, term, input_tuple, assertionString, whereString, swrlString) :
if (pd.notnull(item.Unit)) :
if checkTemplate(item.Unit) :
open_index = item.Unit.find("{")
close_index = item.Unit.find("}")
key = item.Unit[open_index+1:close_index]
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.hasValue + "> " + convertImplicitToKGEntry(key) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["Unit"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["Unit"] + "> " + convertImplicitToKGEntry(key)
whereString += " ;\n <" + properties_tuple["Unit"] + "> ?" + key.lower() + "_E"
swrlString += properties_tuple["Unit"] + "(" + term + " , " + [key,key[1:] + "_V"][checkImplicit(key)] + ") ^ "
input_tuple["Unit"] = key
else :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.hasValue + "> " + str(codeMapper(item.Unit)) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["Unit"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["Unit"] + "> " + str(codeMapper(item.Unit))
whereString += " ;\n <" + properties_tuple["Unit"] + "> " + str(codeMapper(item.Unit))
swrlString += properties_tuple["Unit"] + "(" + term + " , " + str(codeMapper(item.Unit)) + ") ^ "
input_tuple["Unit"] = codeMapper(item.Unit)
# Incorporate item.Format here
return [input_tuple, assertionString, whereString, swrlString]
def writeClassTime(item, term, input_tuple, assertionString, whereString, swrlString) :
if (pd.notnull(item.Time)) :
if checkTemplate(item.Time) :
open_index = item.Time.find("{")
close_index = item.Time.find("}")
key = item.Time[open_index+1:close_index]
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.onProperty + "> <" + properties_tuple["Time"] + "> ;\n <" + owl.someValuesFrom + "> " + convertImplicitToKGEntry(key) + " ]"
#assertionString += " ;\n <" + properties_tuple["Time"] + "> " + convertImplicitToKGEntry(key)
whereString += " ;\n <" + properties_tuple["Time"] + "> ?" + key.lower() + "_E"
swrlString += properties_tuple["Time"] + "(" + term + " , " + [key,key[1:] + "_V"][checkImplicit(key)] + ") ^ "
else :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.onProperty + "> <" + properties_tuple["Time"] + "> ;\n <" + owl.someValuesFrom + "> " + convertImplicitToKGEntry(item.Time) + " ]"
#assertionString += " ;\n <" + properties_tuple["Time"] + "> " + convertImplicitToKGEntry(item.Time)
whereString += " ;\n <" + properties_tuple["Time"] + "> " + [item.Time + " ",item.Time[1:] + "_V "][checkImplicit(item.Time)]
swrlString += properties_tuple["Time"] + "(" + term + " , " + [item.Time + " ",item.Time[1:] + "_V "][checkImplicit(item.Time)] + ") ^ "
input_tuple["Time"]=item.Time
return [input_tuple, assertionString, whereString, swrlString]
def writeClassRelation(item, term, input_tuple, assertionString, whereString, swrlString) :
if (pd.notnull(item.inRelationTo)) :
input_tuple["inRelationTo"]=item.inRelationTo
# If there is a value in the Relation column but not the Role column ...
if (pd.notnull(item.Relation)) and (pd.isnull(item.Role)) :
assertionString += " ;\n " + item.Relation + " " + convertImplicitToKGEntry(item.inRelationTo)
if(isSchemaVar(item.inRelationTo)):
whereString += " ;\n " + item.Relation + " ?" + item.inRelationTo.lower() + "_E "
swrlString += item.Relation + "(" + term + " , " + "?" + item.inRelationTo.lower() + "_E) ^ "
else :
whereString += " ;\n " + item.Relation + " " + [item.inRelationTo + " ",item.inRelationTo[1:] + "_V "][checkImplicit(item.inRelationTo)]
swrlString += item.Relation + "(" + term + " , " + [item.inRelationTo,item.inRelationTo[1:] + "_V"][checkImplicit(item.inRelationTo)] + ") ^ "
input_tuple["Relation"]=item.Relation
# If there is a value in the Role column but not the Relation column ...
elif (pd.isnull(item.Relation)) and (pd.notnull(item.Role)) :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.onProperty + "> <" + properties_tuple["Role"] + "> ;\n <" + owl.someValuesFrom + "> [ <" + rdf.type + "> <" + owl.Class + "> ;\n <" + owl.intersectionOf + "> ( \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + "> " + [item.inRelationTo,convertImplicitToKGEntry(item.inRelationTo)][checkImplicit(item.inRelationTo)] + " ;\n <" + owl.onProperty + "> <" + properties_tuple["inRelationTo"] + "> ] <" + item.Role + "> ) ] ]"
#assertionString += " ;\n <" + properties_tuple["Role"] + "> [ <" + rdf.type + "> " + item.Role + " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(item.inRelationTo) + " ]"
whereString += " ;\n <" + properties_tuple["Role"] + "> [ <" + rdf.type + "> " + item.Role + " ;\n <" + properties_tuple["inRelationTo"] + "> " + [item.inRelationTo + " ",item.inRelationTo[1:] + "_V "][checkImplicit(item.inRelationTo)] + " ]"
swrlString += "" # add appropriate swrl term
input_tuple["Role"]=item.Role
# If there is a value in the Role and Relation columns ...
elif (pd.notnull(item.Relation)) and (pd.notnull(item.Role)) :
input_tuple["Relation"]=item.Relation
input_tuple["Role"]=item.Role
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.onProperty + "> <" + properties_tuple["Role"] + "> ;\n <" + owl.someValuesFrom + "> [ <" + rdf.type + "> <" + owl.Class + "> ;\n <" + owl.intersectionOf + "> ( \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + "> " + [item.inRelationTo,convertImplicitToKGEntry(item.inRelationTo)][checkImplicit(item.inRelationTo)] + " ;\n <" + owl.onProperty + "> <" + item.Relation + "> ] <" + item.Role + "> ) ] ]"
#assertionString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(item.inRelationTo)
if(isSchemaVar(item.inRelationTo)):
whereString += " ;\n <" + properties_tuple["inRelationTo"] + "> ?" + item.inRelationTo.lower() + "_E "
swrlString += "" # add appropriate swrl term
else :
whereString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + [item.inRelationTo + " ",item.inRelationTo[1:] + "_V "][checkImplicit(item.inRelationTo)]
swrlString += "" # add appropriate swrl term
elif (pd.isnull(item.Relation)) and (pd.isnull(item.Role)) :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + "> " + convertImplicitToKGEntry(item.inRelationTo) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["inRelationTo"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(item.inRelationTo)
if(isSchemaVar(item.inRelationTo)):
whereString += " ;\n <" + properties_tuple["inRelationTo"] + "> ?" + item.inRelationTo.lower() + "_E "
swrlString += properties_tuple["inRelationTo"] + "(" + term + " , " + "?" + item.inRelationTo.lower() + "_E) ^ "
else :
whereString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + [item.inRelationTo + " ",item.inRelationTo[1:] + "_V "][checkImplicit(item.inRelationTo)]
swrlString += properties_tuple["inRelationTo"] + "(" + term + " , " + [item.inRelationTo,item.inRelationTo[1:] + "_V"][checkImplicit(item.inRelationTo)] + ") ^ "
elif (pd.notnull(item.Role)) : # if there is a role, but no in relation to
input_tuple["Role"]=item.Role
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.onProperty + "> <" + properties_tuple["Role"] + "> ;\n <" + owl.someValuesFrom + "> [ <" + rdf.type + "> <" + item.Role + "> ] ]"
#assertionString += " ;\n <" + properties_tuple["Role"] + "> [ <" + rdf.type + "> " + item.Role + " ]"
whereString += " ;\n <" + properties_tuple["Role"] + "> [ <" + rdf.type + "> " + item.Role + " ]"
swrlString += "" # add appropriate swrl term
return [input_tuple, assertionString, whereString, swrlString]
def writeClassWasDerivedFrom(item, term, input_tuple, provenanceString, whereString, swrlString) :
if pd.notnull(item.wasDerivedFrom) :
provenanceString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.someValuesFrom + "> " + convertImplicitToKGEntry(item.wasDerivedFrom) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["wasDerivedFrom"] + "> ]"
#provenanceString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> " + convertImplicitToKGEntry(item.wasDerivedFrom)
input_tuple["wasDerivedFrom"]=item.wasDerivedFrom
if(isSchemaVar(item.wasDerivedFrom)):
whereString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> ?" + item.wasDerivedFrom.lower() + "_E "
swrlString += properties_tuple["wasDerivedFrom"] + "(" + term + " , " + "?" + item.wasDerivedFrom.lower() + "_E) ^ "
elif checkTemplate(item.wasDerivedFrom) :
open_index = item.wasDerivedFrom.find("{")
close_index = item.wasDerivedFrom.find("}")
key = item.wasDerivedFrom[open_index+1:close_index]
provenanceString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.someValuesFrom + "> " + convertImplicitToKGEntry(key) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["wasDerivedFrom"] + "> ]"
#provenanceString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> " + convertImplicitToKGEntry(key)
whereString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> ?" + key.lower() + "_E"
swrlString += properties_tuple["wasDerivedFrom"] + "(" + term + " , " + [key,key[1:] + "_V"][checkImplicit(key)] + ") ^ "
else :
whereString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> " + [item.wasDerivedFrom + " ",item.wasDerivedFrom[1:] + "_V "][checkImplicit(item.wasDerivedFrom)]
swrlString += properties_tuple["wasDerivedFrom"] + "(" + term + " , " + [item.wasDerivedFrom,item.wasDerivedFrom[1:] + "_V"][checkImplicit(item.wasDerivedFrom)] + ") ^ "
return [input_tuple, provenanceString, whereString, swrlString]
def writeClassWasGeneratedBy(item, term, input_tuple, provenanceString, whereString, swrlString) :
if pd.notnull(item.wasGeneratedBy) :
provenanceString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> " + convertImplicitToKGEntry(item.wasGeneratedBy)
input_tuple["wasGeneratedBy"]=item.wasGeneratedBy
if(isSchemaVar(item.wasGeneratedBy)):
whereString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> ?" + item.wasGeneratedBy.lower() + "_E "
swrlString += properties_tuple["wasGeneratedBy"] + "(" + term + " , " + "?" + item.wasGeneratedBy.lower() + "_E) ^ "
elif checkTemplate(item.wasGeneratedBy) :
open_index = item.wasGeneratedBy.find("{")
close_index = item.wasGeneratedBy.find("}")
key = item.wasGeneratedBy[open_index+1:close_index]
assertionString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> " + convertImplicitToKGEntry(key)
whereString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> ?" + key.lower() + "_E"
swrlString += properties_tuple["wasGeneratedBy"] + "(" + term + " , " + [key,key[1:] + "_V"][checkImplicit(key)] + ") ^ "
else :
whereString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> " + [item.wasGeneratedBy + " ",item.wasGeneratedBy[1:] + "_V "][checkImplicit(item.wasGeneratedBy)]
swrlString += properties_tuple["wasGeneratedBy"] + "(" + term + " , " + [item.wasGeneratedBy,item.wasGeneratedBy[1:] + "_V"][checkImplicit(item.wasGeneratedBy)] + ") ^ "
return [input_tuple, provenanceString, whereString, swrlString]
def writeImplicitEntryTuples(implicit_entry_list, timeline_tuple, output_file, query_file, swrl_file, dm_fn) :
implicit_entry_tuples = []
assertionString = ''
provenanceString = ''
whereString = '\n'
swrlString = ''
datasetIdentifier = hashlib.md5(dm_fn.encode('utf-8')).hexdigest()
output_file.write("<" + prefixes[kb] + "head-implicit_entry-" + datasetIdentifier + "> { ")
output_file.write("\n <" + prefixes[kb] + "nanoPub-implicit_entry-" + datasetIdentifier + "> <" + rdf.type + "> <" + np.Nanopublication + ">")
output_file.write(" ;\n <" + np.hasAssertion + "> <" + prefixes[kb] + "assertion-implicit_entry-" + datasetIdentifier + ">")
output_file.write(" ;\n <" + np.hasProvenance + "> <" + prefixes[kb] + "provenance-implicit_entry-" + datasetIdentifier + ">")
output_file.write(" ;\n <" + np.hasPublicationInfo + "> <" + prefixes[kb] + "pubInfo-implicit_entry-" + datasetIdentifier + ">")
output_file.write(" .\n}\n\n")
col_headers=list(pd.read_csv(dm_fn).columns.values)
for item in implicit_entry_list :
implicit_tuple = {}
if "Template" in col_headers and pd.notnull(item.Template) :
implicit_tuple["Template"]=item.Template
assertionString += "\n <" + prefixes[kb] + item.Column[2:] + "> <" + rdf.type + "> owl:Class"
term_implicit = item.Column[1:] + "_V"
whereString += " " + term_implicit + " <" + rdf.type + "> "
implicit_tuple["Column"]=item.Column
if (hasattr(item,"Label") and pd.notnull(item.Label)) :
implicit_tuple["Label"]=item.Label
if ',' in item.Label :
labels = parseString(item.Label,',')
for label in labels :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + label + "\"^^xsd:string"
else :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + item.Label + "\"^^xsd:string"
else :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + item.Column[2:] + "\"^^xsd:string"
implicit_tuple["Label"]=item.Column[2:]
if (hasattr(item,"Comment") and pd.notnull(item.Comment)) :
assertionString += " ;\n <" + properties_tuple["Comment"] + "> \"" + item.Comment + "\"^^xsd:string"
implicit_tuple["Comment"]=item.Comment
[implicit_tuple, assertionString, whereString, swrlString] = writeClassAttributeOrEntity(item, term_implicit, implicit_tuple, assertionString, whereString, swrlString)
[implicit_tuple, assertionString, whereString, swrlString] = writeClassAttributeOf(item, term_implicit, implicit_tuple, assertionString, whereString, swrlString)
[implicit_tuple, assertionString, whereString, swrlString] = writeClassUnit(item, term_implicit, implicit_tuple, assertionString, whereString, swrlString)
[implicit_tuple, assertionString, whereString, swrlString] = writeClassTime(item, term_implicit, implicit_tuple, assertionString, whereString, swrlString)
[implicit_tuple, assertionString, whereString, swrlString] = writeClassRelation(item, term_implicit, implicit_tuple, assertionString, whereString, swrlString)
assertionString += " .\n"
provenanceString += "\n <" + prefixes[kb] + item.Column[2:] + ">"
provenanceString +="\n <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime"
[implicit_tuple, provenanceString, whereString, swrlString] = writeClassWasGeneratedBy(item, term_implicit, implicit_tuple, provenanceString, whereString, swrlString)
[implicit_tuple, provenanceString, whereString, swrlString] = writeClassWasDerivedFrom(item, term_implicit, implicit_tuple, provenanceString, whereString, swrlString)
provenanceString += " .\n"
whereString += ".\n\n"
implicit_entry_tuples.append(implicit_tuple)
if timeline_tuple != {}:
for key in timeline_tuple :
assertionString += "\n " + convertImplicitToKGEntry(key) + " <" + rdf.type + "> owl:Class "
for timeEntry in timeline_tuple[key] :
if 'Type' in timeEntry :
assertionString += " ;\n rdfs:subClassOf " + timeEntry['Type']
if 'Label' in timeEntry :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + timeEntry['Label'] + "\"^^xsd:string"
if 'Start' in timeEntry and 'End' in timeEntry and timeEntry['Start'] == timeEntry['End']:
assertionString += " ;\n <" + sio.hasValue + "> " + str(timeEntry['Start']) # rewrite this as a restriction
if 'Start' in timeEntry :
if 'Unit' in timeEntry :
assertionString += " ;\n <" + rdfs.subClassOf + ">\n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + ">\n [ <" + rdf.type + "> <" + owl.Class + "> ;\n <" + owl.intersectionOf + "> ( [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.hasValue + "> " + str(timeEntry['Start']) +" ;\n <" + owl.onProperty + "> <" + sio.hasValue + "> ] " + str(codeMapper(timeEntry['Unit'])) + " ) ] ;\n <" + owl.onProperty + "> <" + properties_tuple["Start"] + "> ] "
else : # update restriction that gets generated if unit is not specified
assertionString += " ;\n <" + properties_tuple["Start"] + "> [ <" + sio.hasValue + "> " + str(timeEntry['Start']) + " ]"
if 'End' in timeEntry :
if 'Unit' in timeEntry :
assertionString += " ;\n <" + rdfs.subClassOf + ">\n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.allValuesFrom + ">\n [ <" + rdf.type + "> <" + owl.Class + "> ;\n <" + owl.intersectionOf + "> ( [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.hasValue + "> " + str(timeEntry['End']) +" ;\n <" + owl.onProperty + "> <" + sio.hasValue + "> ] " + str(codeMapper(timeEntry['Unit'])) + " ) ] ;\n <" + owl.onProperty + "> <" + properties_tuple["End"] + "> ] "
else : # update restriction that gets generated if unit is not specified
assertionString += " ;\n <" + properties_tuple["End"] + "> [ <" + sio.hasValue + "> " + str(timeEntry['End']) + " ]"
if 'Unit' in timeEntry :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.hasValue + "> " + str(codeMapper(timeEntry['Unit'])) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["Unit"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["Unit"] + "> " + timeEntry['Unit']
if 'inRelationTo' in timeEntry :
assertionString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(timeEntry['inRelationTo'])
assertionString += " .\n"
provenanceString += "\n " + convertImplicitToKGEntry(key) + " <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .\n"
output_file.write("<" + prefixes[kb] + "assertion-implicit_entry-" + datasetIdentifier + "> {")
output_file.write(assertionString + "\n}\n\n")
output_file.write("<" + prefixes[kb] + "provenance-implicit_entry-" + datasetIdentifier + "> {")
provenanceString = "\n <" + prefixes[kb] + "assertion-implicit_entry-" + datasetIdentifier + "> <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .\n" + provenanceString
output_file.write(provenanceString + "\n}\n\n")
output_file.write("<" + prefixes[kb] + "pubInfo-implicit_entry-" + datasetIdentifier + "> {\n <" + prefixes[kb] + "nanoPub-implicit_entry-" + datasetIdentifier + "> <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .\n}\n\n")
whereString += "}"
#print(whereString)
query_file.write(whereString)
swrl_file.write(swrlString[:-2])
return implicit_entry_tuples
def writeExplicitEntryTuples(explicit_entry_list, output_file, query_file, swrl_file, dm_fn) :
explicit_entry_tuples = []
assertionString = ''
provenanceString = ''
publicationInfoString = ''
selectString = "SELECT DISTINCT "
whereString = "WHERE {\n"
swrlString = ""
datasetIdentifier = hashlib.md5(dm_fn.encode('utf-8')).hexdigest()
output_file.write("<" + prefixes[kb] + "head-explicit_entry-" + datasetIdentifier + "> { ")
output_file.write("\n <" + prefixes[kb] + "nanoPub-explicit_entry-" + datasetIdentifier + "> <" + rdf.type + "> <" + np.Nanopublication + ">")
output_file.write(" ;\n <" + np.hasAssertion + "> <" + prefixes[kb] + "assertion-explicit_entry-" + datasetIdentifier + ">")
output_file.write(" ;\n <" + np.hasProvenance + "> <" + prefixes[kb] + "provenance-explicit_entry-" + datasetIdentifier + ">")
output_file.write(" ;\n <" + np.hasPublicationInfo + "> <" + prefixes[kb] + "pubInfo-explicit_entry-" + datasetIdentifier + ">")
output_file.write(" .\n}\n\n")
col_headers=list(pd.read_csv(dm_fn).columns.values)
for item in explicit_entry_list :
explicit_entry_tuple = {}
if "Template" in col_headers and pd.notnull(item.Template) :
explicit_entry_tuple["Template"]=item.Template
term = item.Column.replace(" ","_").replace(",","").replace("(","").replace(")","").replace("/","-").replace("\\","-")
assertionString += "\n <" + prefixes[kb] + term + "> <" + rdf.type + "> owl:Class"
selectString += "?" + term.lower() + " "
whereString += " ?" + term.lower() + "_E <" + rdf.type + "> "
term_expl = "?" + term.lower() + "_E"
#print(item.Column
explicit_entry_tuple["Column"]=item.Column
[explicit_entry_tuple, assertionString, whereString, swrlString] = writeClassAttributeOrEntity(item, term_expl, explicit_entry_tuple, assertionString, whereString, swrlString)
[explicit_entry_tuple, assertionString, whereString, swrlString] = writeClassAttributeOf(item, term_expl, explicit_entry_tuple, assertionString, whereString, swrlString)
[explicit_entry_tuple, assertionString, whereString, swrlString] = writeClassUnit(item, term_expl, explicit_entry_tuple, assertionString, whereString, swrlString)
[explicit_entry_tuple, assertionString, whereString, swrlString] = writeClassTime(item, term_expl, explicit_entry_tuple, assertionString, whereString, swrlString)
[explicit_entry_tuple, assertionString, whereString, swrlString] = writeClassRelation(item, term_expl, explicit_entry_tuple, assertionString, whereString, swrlString)
if "Label" in col_headers and (pd.notnull(item.Label)) :
if ',' in item.Label :
labels = parseString(item.Label,',')
for label in labels :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + label + "\"^^xsd:string"
else :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + item.Label + "\"^^xsd:string"
explicit_entry_tuple["Label"]=item.Label
if "Comment" in col_headers and (pd.notnull(item.Comment)) :
assertionString += " ;\n <" + properties_tuple["Comment"] + "> \"" + item.Comment + "\"^^xsd:string"
explicit_entry_tuple["Comment"]=item.Comment
assertionString += " .\n"
provenanceString += "\n <" + prefixes[kb] + term + ">"
provenanceString += "\n <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime"
[explicit_entry_tuple, provenanceString, whereString, swrlString] = writeClassWasGeneratedBy(item, term_expl, explicit_entry_tuple, provenanceString, whereString, swrlString)
[explicit_entry_tuple, provenanceString, whereString, swrlString] = writeClassWasDerivedFrom(item, term_expl, explicit_entry_tuple, provenanceString, whereString, swrlString)
provenanceString += " .\n"
whereString += " ;\n <" + sio.hasValue + "> ?" + term.lower() + " .\n\n"
if "hasPosition" in col_headers and pd.notnull(item.hasPosition) :
publicationInfoString += "\n <" + prefixes[kb] + term + "> hasco:hasPosition \"" + str(item.hasPosition) + "\"^^xsd:integer ."
explicit_entry_tuple["hasPosition"]=item.hasPosition
explicit_entry_tuples.append(explicit_entry_tuple)
output_file.write("<" + prefixes[kb] + "assertion-explicit_entry-" + datasetIdentifier + "> {")
output_file.write(assertionString + "\n}\n\n")
output_file.write("<" + prefixes[kb] + "provenance-explicit_entry-" + datasetIdentifier + "> {")
provenanceString = "\n <" + prefixes[kb] + "assertion-explicit_entry-" + datasetIdentifier + "> <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .\n" + provenanceString
output_file.write(provenanceString + "\n}\n\n")
output_file.write("<" + prefixes[kb] + "pubInfo-explicit_entry-" + datasetIdentifier + "> {\n <" + prefixes[kb] + "nanoPub-explicit_entry-" + datasetIdentifier + "> <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .")
output_file.write(publicationInfoString + "\n}\n\n")
#print(selectString)
#print(whereString)
query_file.write(selectString)
query_file.write(whereString)
swrl_file.write(swrlString)
return explicit_entry_tuples
def writeImplicitEntry(assertionString, provenanceString,publicationInfoString, explicit_entry_tuples, implicit_entry_tuples, timeline_tuple, vref_list, v_column, index, row, col_headers) :
try :
#col_headers=list(pd.read_csv(dm_fn).columns.values)
if timeline_tuple != {} :
if v_column in timeline_tuple :
v_id = hashlib.md5((str(timeline_tuple[v_column]) + str(index)).encode("utf-8")).hexdigest()
assertionString += "\n " + convertImplicitToKGEntry(v_column, v_id) + " <" + rdf.type + "> " + convertImplicitToKGEntry(v_column)
for timeEntry in timeline_tuple[v_column] :
if 'Type' in timeEntry :
assertionString += " ;\n <" + rdf.type + "> " + timeEntry['Type']
if 'Label' in timeEntry :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + timeEntry['Label'] + "\"^^xsd:string"
if 'Start' in timeEntry and 'End' in timeEntry and timeEntry['Start'] == timeEntry['End']:
assertionString += " ;\n <" + sio.hasValue + "> " + str(timeEntry['Start'])
if 'Start' in timeEntry :
assertionString += " ;\n <" + properties_tuple["Start"] + "> [ <" + sio.hasValue + "> " + str(timeEntry['Start']) + " ]"
if 'End' in timeEntry :
assertionString += " ;\n <" + properties_tuple["End"] + "> [ <" + sio.hasValue + "> " + str(timeEntry['End']) + " ]"
if 'Unit' in timeEntry :
assertionString += " ;\n <" + rdfs.subClassOf + "> \n [ <" + rdf.type + "> <" + owl.Restriction + "> ;\n <" + owl.hasValue + "> " + str(codeMapper(timeEntry['Unit'])) + " ;\n <" + owl.onProperty + "> <" + properties_tuple["Unit"] + "> ]"
#assertionString += " ;\n <" + properties_tuple["Unit"] + "> " + timeEntry['Unit']
if 'inRelationTo' in timeEntry :
assertionString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(timeEntry['inRelationTo'], v_id)
if checkImplicit(timeEntry['inRelationTo']) and timeEntry['inRelationTo'] not in vref_list :
vref_list.append(timeEntry['inRelationTo'])
assertionString += " .\n"
for v_tuple in implicit_entry_tuples :
if (v_tuple["Column"] == v_column) :
if "Study" in v_tuple :
continue
else :
v_id = hashlib.md5((str(v_tuple) + str(index)).encode("utf-8")).hexdigest()
if "Template" in v_tuple :
template_term = extractTemplate(col_headers,row,v_tuple["Template"])
termURI = "<" + prefixes[kb] + template_term + ">"
else :
termURI = "<" + prefixes[kb] + v_tuple["Column"][2:] + "-" + v_id + ">"
assertionString += "\n " + termURI + " <" + rdf.type + "> <" + prefixes[kb] + v_tuple["Column"][2:] + ">"
if "Entity" in v_tuple :
if ',' in v_tuple["Entity"] :
entities = parseString(v_tuple["Entity"],',')
for entity in entities :
assertionString += " ;\n <" + rdf.type + "> " + entity
else :
assertionString += " ;\n <" + rdf.type + "> " + v_tuple["Entity"]
if "Attribute" in v_tuple :
if ',' in v_tuple["Attribute"] :
attributes = parseString(v_tuple["Attribute"],',')
for attribute in attributes :
assertionString += " ;\n <" + rdf.type + "> " + attribute
else :
assertionString += " ;\n <" + rdf.type + "> " + v_tuple["Attribute"]
# Need to get the right ID uri if we put this in.. Commenting out identifier for now
#if "Subject" in v_tuple :
# assertionString += " ;\n sio:hasIdentifier <" + prefixes[kb] + v_tuple["Subject"] + "-" + v_id + ">" #should be actual ID
if "Label" in v_tuple :
if ',' in v_tuple["Label"] :
labels = parseString(v_tuple["Label"],',')
for label in labels :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + label + "\"^^xsd:string"
else :
assertionString += " ;\n <" + properties_tuple["Label"] + "> \"" + v_tuple["Label"] + "\"^^xsd:string"
if "Time" in v_tuple :
if checkImplicit(v_tuple["Time"]) :
for vr_tuple in implicit_entry_tuples :
if (vr_tuple["Column"] == v_tuple["Time"]) :
timeID = hashlib.md5((str(vr_tuple) + str(index)).encode("utf-8")).hexdigest()
assertionString += " ;\n <" + properties_tuple["Time"] + "> " + convertImplicitToKGEntry(v_tuple["Time"], timeID)
if v_tuple["Time"] not in vref_list :
vref_list.append(v_tuple["Time"])
else :
assertionString += " ;\n <" + properties_tuple["Time"] + "> " + convertImplicitToKGEntry(v_tuple["Time"], v_id) #should be actual ID
if "inRelationTo" in v_tuple :
relationToID = None
for vr_tuple in implicit_entry_tuples :
if (vr_tuple["Column"] == v_tuple["inRelationTo"]) :
relationToID = hashlib.md5((str(vr_tuple) + str(index)).encode("utf-8")).hexdigest()
if ("Role" in v_tuple) and ("Relation" not in v_tuple) :
assertionString += " ;\n <" + properties_tuple["Role"] + "> [ <" + rdf.type + "> " + v_tuple["Role"] + " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(v_tuple["inRelationTo"], relationToID) + " ]"
elif ("Role" not in v_tuple) and ("Relation" in v_tuple) :
assertionString += " ;\n " + v_tuple["Relation"] + " " + convertImplicitToKGEntry(v_tuple["inRelationTo"],v_id)
assertionString += " ;\n " + v_tuple["Relation"] + " " + convertImplicitToKGEntry(v_tuple["inRelationTo"],relationToID)
elif ("Role" not in v_tuple) and ("Relation" not in v_tuple) :
assertionString += " ;\n <" + properties_tuple["inRelationTo"] + "> " + convertImplicitToKGEntry(v_tuple["inRelationTo"],relationToID)
elif "Role" in v_tuple :
assertionString += " ;\n <" + properties_tuple["Role"] + "> [ <" + rdf.type + "> " + v_tuple["Role"] + " ]"
assertionString += " .\n"
provenanceString += "\n " + termURI + " <" + prov.generatedAtTime + "> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime"
if "wasGeneratedBy" in v_tuple :
if ',' in v_tuple["wasGeneratedBy"] :
generatedByTerms = parseString(v_tuple["wasGeneratedBy"],',')
for generatedByTerm in generatedByTerms :
provenanceString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> " + convertImplicitToKGEntry(generatedByTerm,v_id)
if checkImplicit(generatedByTerm) and generatedByTerm not in vref_list :
vref_list.append(generatedByTerm)
else :
provenanceString += " ;\n <" + properties_tuple["wasGeneratedBy"] + "> " + convertImplicitToKGEntry(v_tuple["wasGeneratedBy"],v_id)
if checkImplicit(v_tuple["wasGeneratedBy"]) and v_tuple["wasGeneratedBy"] not in vref_list :
vref_list.append(v_tuple["wasGeneratedBy"]);
if "wasDerivedFrom" in v_tuple :
if ',' in v_tuple["wasDerivedFrom"] :
derivedFromTerms = parseString(v_tuple["wasDerivedFrom"],',')
for derivedFromTerm in derivedFromTerms :
provenanceString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> " + convertImplicitToKGEntry(derivedFromTerm,v_id)
if checkImplicit(derivedFromTerm) and derivedFromTerm not in vref_list :
vref_list.append(derivedFromTerm);
else :
provenanceString += " ;\n <" + properties_tuple["wasDerivedFrom"] + "> " + convertImplicitToKGEntry(v_tuple["wasDerivedFrom"],v_id)
if checkImplicit(v_tuple["wasDerivedFrom"]) and v_tuple["wasDerivedFrom"] not in vref_list :
vref_list.append(v_tuple["wasDerivedFrom"]);
#if "wasGeneratedBy" in v_tuple or "wasDerivedFrom" in v_tuple :
provenanceString += " .\n"
return [assertionString,provenanceString,publicationInfoString,vref_list]
except Exception as e :
print("Warning: Unable to create implicit entry: " + str(e))
def processInfosheet(output_file, dm_fn, cb_fn, cmap_fn, timeline_fn):
infosheet_tuple = {}
if 'infosheet' in config['Source Files'] :
infosheet_fn = config['Source Files']['infosheet']
try :
infosheet_file = pd.read_csv(infosheet_fn, dtype=object)
except Exception as e :
print("Warning: Collection metadata will not be written to the output file.\nThe specified Infosheet file does not exist or is unreadable: " + str(e))
return [dm_fn, cb_fn, cmap_fn, timeline_fn]
for row in infosheet_file.itertuples() :
if(pd.notnull(row.Value)):
infosheet_tuple[row.Attribute]=row.Value
# If SDD files included in Infosheet, they override the config declarations
if "Dictionary Mapping" in infosheet_tuple :
dm_fn = infosheet_tuple["Dictionary Mapping"]
if "Codebook" in infosheet_tuple :
cb_fn = infosheet_tuple["Codebook"]
if "Code Mapping" in infosheet_tuple :
cmap_fn = infosheet_tuple["Code Mapping"]
if "Timeline" in infosheet_tuple :
timeline_fn = infosheet_tuple["Timeline"]
datasetIdentifier = hashlib.md5(dm_fn.encode('utf-8')).hexdigest()
output_file.write("<" + prefixes[kb] + "head-collection_metadata-" + datasetIdentifier + "> { ")
output_file.write("\n <" + prefixes[kb] + "nanoPub-collection_metadata-" + datasetIdentifier + "> <" + rdf.type + "> <" + np.Nanopublication + ">")
output_file.write(" ;\n <" + np.hasAssertion + "> <" + prefixes[kb] + "assertion-collection_metadata-" + datasetIdentifier + ">")
output_file.write(" ;\n <" + np.hasProvenance + "> <" + prefixes[kb] + "provenance-collection_metadata-" + datasetIdentifier + ">")
output_file.write(" ;\n <" + np.hasPublicationInfo + "> <" + prefixes[kb] + "pubInfo-collection_metadata-" + datasetIdentifier + ">")
output_file.write(" .\n}\n\n")
assertionString = "<" + prefixes[kb] + "collection-" + datasetIdentifier + ">"
provenanceString = " <" + prefixes[kb] + "collection-" + datasetIdentifier + "> <http://www.w3.org/ns/prov#generatedAtTime> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime"
if "Type" in infosheet_tuple :
assertionString += " <" + rdf.type + "> " + [infosheet_tuple["Type"],"<" + infosheet_tuple["Type"] + ">"][isURI(infosheet_tuple["Type"])]
else :
assertionString += " <" + rdf.type + "> <http://purl.org/dc/dcmitype/Collection>"
#print("Warning: The Infosheet file is missing the Type value declaration")
#sys.exit(1)
if "Title" in infosheet_tuple :
assertionString += " ;\n <http://purl.org/dc/terms/title> \"" + infosheet_tuple["Title"] + "\"^^xsd:string"
if "Alternative Title" in infosheet_tuple :
if ',' in infosheet_tuple["Alternative Title"] :
alt_titles = parseString(infosheet_tuple["Alternative Title"],',')
for alt_title in alt_titles :
assertionString += " ;\n <http://purl.org/dc/terms/alternative> \"" + alt_title + "\"^^xsd:string"
else :
assertionString += " ;\n <http://purl.org/dc/terms/alternative> \"" + infosheet_tuple["Alternative Title"] + "\"^^xsd:string"
if "Comment" in infosheet_tuple :
assertionString += " ;\n <http://www.w3.org/2000/01/rdf-schema#comment> \"" + infosheet_tuple["Comment"] + "\"^^xsd:string"
if "Description" in infosheet_tuple :
assertionString += " ;\n <http://purl.org/dc/terms/description> \"" + infosheet_tuple["Description"] + "\"^^xsd:string"
if "Date Created" in infosheet_tuple :
provenanceString += " ;\n <http://purl.org/dc/terms/created> \"" + infosheet_tuple["Date Created"] + "\"^^xsd:date"
if "Creators" in infosheet_tuple :
if ',' in infosheet_tuple["Creators"] :
creators = parseString(infosheet_tuple["Creators"],',')
for creator in creators :
provenanceString += " ;\n <http://purl.org/dc/terms/creator> " + ["\"" + creator + "\"^^xsd:string","<" + creator + ">"][isURI(creator)]
else :
provenanceString += " ;\n <http://purl.org/dc/terms/creator> " + ["\"" + infosheet_tuple["Creators"] + "\"^^xsd:string","<" + infosheet_tuple["Creators"] + ">"][isURI(infosheet_tuple["Creators"])]
if "Contributors" in infosheet_tuple :
if ',' in infosheet_tuple["Contributors"] :
contributors = parseString(infosheet_tuple["Contributors"],',')
for contributor in contributors :
provenanceString += " ;\n <http://purl.org/dc/terms/contributor> " + ["\"" + contributor + "\"^^xsd:string","<" + contributor + ">"][isURI(contributor)]
else :
provenanceString += " ;\n <http://purl.org/dc/terms/contributor> " + ["\"" + infosheet_tuple["Contributors"] + "\"^^xsd:string","<" + infosheet_tuple["Contributors"] + ">"][isURI(infosheet_tuple["Contributors"])]
if "Publisher" in infosheet_tuple :
if ',' in infosheet_tuple["Publisher"] :
publishers = parseString(infosheet_tuple["Publisher"],',')
for publisher in publishers :
provenanceString += " ;\n <http://purl.org/dc/terms/publisher> " + ["\"" + publisher + "\"^^xsd:string","<" + publisher + ">"][isURI(publisher)]
else :
provenanceString += " ;\n <http://purl.org/dc/terms/publisher> " + ["\"" + infosheet_tuple["Publisher"] + "\"^^xsd:string","<" + infosheet_tuple["Publisher"] + ">"][isURI(infosheet_tuple["Publisher"])]
if "Date of Issue" in infosheet_tuple :
provenanceString += " ;\n <http://purl.org/dc/terms/issued> \"" + infosheet_tuple["Date of Issue"] + "\"^^xsd:date"
if "Link" in infosheet_tuple :
assertionString += " ;\n <http://xmlns.com/foaf/0.1/page> <" + infosheet_tuple["Link"] + ">"
if "Identifier" in infosheet_tuple :
assertionString += " ;\n <http://semanticscience.org/resource/hasIdentifier> \n [ <" + rdf.type + "> <http://semanticscience.org/resource/Identifier> ; \n <http://semanticscience.org/resource/hasValue> \"" + infosheet_tuple["Identifier"] + "\"^^xsd:string ]"
if "Keywords" in infosheet_tuple :
if ',' in infosheet_tuple["Keywords"] :
keywords = parseString(infosheet_tuple["Keywords"],',')
for keyword in keywords :
assertionString += " ;\n <http://www.w3.org/ns/dcat#keyword> \"" + keyword + "\"^^xsd:string"
else :
assertionString += " ;\n <http://www.w3.org/ns/dcat#keyword> \"" + infosheet_tuple["Keywords"] + "\"^^xsd:string"
if "License" in infosheet_tuple :
if ',' in infosheet_tuple["License"] :
licenses = parseString(infosheet_tuple["License"],',')
for license in licenses :
assertionString += " ;\n <http://purl.org/dc/terms/license> " + ["\"" + license + "\"^^xsd:string","<" + license + ">"][isURI(license)]
else :
assertionString += " ;\n <http://purl.org/dc/terms/license> " + ["\"" + infosheet_tuple["License"] + "\"^^xsd:string","<" + infosheet_tuple["License"] + ">"][isURI(infosheet_tuple["License"])]
if "Rights" in infosheet_tuple :
if ',' in infosheet_tuple["Rights"] :
rights = parseString(infosheet_tuple["Rights"],',')
for right in rights :
assertionString += " ;\n <http://purl.org/dc/terms/rights> " + ["\"" + right + "\"^^xsd:string","<" + right + ">"][isURI(right)]
else :
assertionString += " ;\n <http://purl.org/dc/terms/rights> " + ["\"" + infosheet_tuple["Rights"] + "\"^^xsd:string","<" + infosheet_tuple["Rights"] + ">"][isURI(infosheet_tuple["Rights"])]
if "Language" in infosheet_tuple :
assertionString += " ;\n <http://purl.org/dc/terms/language> \"" + infosheet_tuple["Language"] + "\"^^xsd:string"
if "Version" in infosheet_tuple :
provenanceString += " ;\n <http://purl.org/pav/version> " + ["\"" + infosheet_tuple["Version"] + "\"^^xsd:string","<" + infosheet_tuple["Version"] + ">"][isURI(infosheet_tuple["Version"])]
provenanceString += " ;\n <http://www.w3.org/2002/07/owl/versionInfo> " + ["\"" + infosheet_tuple["Version"] + "\"^^xsd:string","<" + infosheet_tuple["Version"] + ">"][isURI(infosheet_tuple["Version"])]
if "Previous Version" in infosheet_tuple :
provenanceString += " ;\n <http://purl.org/pav/previousVersion> " + ["\"" + infosheet_tuple["Previous Version"] + "\"^^xsd:string","<" + infosheet_tuple["Previous Version"] + ">"][isURI(infosheet_tuple["Previous Version"])]
if "Version Of" in infosheet_tuple :
provenanceString += " ;\n <http://purl.org/dc/terms/isVersionOf> " + ["\"" + infosheet_tuple["Version Of"] + "\"^^xsd:string","<" + infosheet_tuple["Version Of"] + ">"][isURI(infosheet_tuple["Version Of"])]
if "Standards" in infosheet_tuple :
if ',' in infosheet_tuple["Standards"] :
standards = parseString(infosheet_tuple["Standards"],',')
for standard in standards :
assertionString += " ;\n <http://purl.org/dc/terms/conformsTo> " + ["\"" + standard + "\"^^xsd:string","<" + standard + ">"][isURI(standard)]
else :
assertionString += " ;\n <http://purl.org/dc/terms/conformsTo> " + ["\"" + infosheet_tuple["Standards"] + "\"^^xsd:string","<" + infosheet_tuple["Standards"] + ">"][isURI(infosheet_tuple["Standards"])]
if "Source" in infosheet_tuple :
if ',' in infosheet_tuple["Source"] :
sources = parseString(infosheet_tuple["Source"],',')
for source in sources :
provenanceString += " ;\n <http://purl.org/dc/terms/source> \"" + source + "\"^^xsd:string"
else :
provenanceString += " ;\n <http://purl.org/dc/terms/source> " + ["\"" + infosheet_tuple["Source"] + "\"^^xsd:string","<" + infosheet_tuple["Source"] + ">"][isURI(infosheet_tuple["Source"])]
if "File Format" in infosheet_tuple :
assertionString += " ;\n <http://purl.org/dc/terms/format> \"" + infosheet_tuple["File Format"] + "\"^^xsd:string"
if "Documentation" in infosheet_tuple : # currently encoded as URI, should confirm that it really is one
provenanceString += " ;\n <http://www.w3.org/ns/dcat#landingPage> <" + infosheet_tuple["Documentation"] + ">"
if "Imports" in infosheet_tuple :
if ',' in infosheet_tuple["Imports"] :
imports = parseString(infosheet_tuple["Imports"],',')
for imp in imports :
assertionString += " ;\n <http://www.w3.org/2002/07/owl#imports> " + [imp,"<" + imp + ">"][isURI(imp)]
else :
assertionString += " ;\n <http://www.w3.org/2002/07/owl#imports> " + [infosheet_tuple["Imports"],"<" + infosheet_tuple["Imports"] + ">"][isURI(infosheet_tuple["Imports"])]
assertionString += " .\n"
provenanceString += " .\n"
output_file.write("<" + prefixes[kb] + "assertion-collection_metadata-" + datasetIdentifier + "> {\n " + assertionString + "\n}\n\n")
output_file.write("<" + prefixes[kb] + "provenance-collection_metadata-" + datasetIdentifier + "> {\n <" + prefixes[kb] + "assertion-dataset_metadata-" + datasetIdentifier + "> <http://www.w3.org/ns/prov#generatedAtTime> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .\n" + provenanceString + "\n}\n\n")
output_file.write("<" + prefixes[kb] + "pubInfo-collection_metadata-" + datasetIdentifier + "> {")
publicationInfoString = "\n <" + prefixes[kb] + "nanoPub-collection_metadata-" + datasetIdentifier + "> <http://www.w3.org/ns/prov#generatedAtTime> \"" + "{:4d}-{:02d}-{:02d}".format(datetime.utcnow().year,datetime.utcnow().month,datetime.utcnow().day) + "T" + "{:02d}:{:02d}:{:02d}".format(datetime.utcnow().hour,datetime.utcnow().minute,datetime.utcnow().second) + "Z\"^^xsd:dateTime .\n"
output_file.write(publicationInfoString + "\n}\n\n")
return [dm_fn, cb_fn, cmap_fn, timeline_fn]
def processPrefixes(output_file,query_file):
prefixes = {}
if 'prefixes' in config['Prefixes']:
prefix_fn = config['Prefixes']['prefixes']
else:
prefix_fn="prefixes.csv"
try:
prefix_file = pd.read_csv(prefix_fn, dtype=object)
for row in prefix_file.itertuples() :
prefixes[row.prefix] = row.url
for prefix in prefixes :
#print(prefix.find(">"))
output_file.write("@prefix " + prefix + ": <" + prefixes[prefix] + "> .\n")
query_file.write("prefix " + prefix + ": <" + prefixes[prefix] + "> \n")
query_file.write("\n")
output_file.write("\n")
except Exception as e :
print("Warning: Something went wrong when trying to read the prefixes file: " + str(e))
return prefixes
def processCodeMappings(cmap_fn):
unit_code_list = []
unit_uri_list = []
unit_label_list = []
if cmap_fn is not None :
try :
code_mappings_reader = pd.read_csv(cmap_fn)
#Using itertuples on a data frame makes the column heads case-sensitive
for code_row in code_mappings_reader.itertuples() :
if pd.notnull(code_row.code):
unit_code_list.append(code_row.code)
if pd.notnull(code_row.uri):
unit_uri_list.append(code_row.uri)
if pd.notnull(code_row.label):
unit_label_list.append(code_row.label)
except Exception as e :
print("Warning: Something went wrong when trying to read the Code Mappings file: " + str(e))
return [unit_code_list,unit_uri_list,unit_label_list]
def processProperties():
properties_tuple = {'Comment': rdfs.comment, 'attributeOf': sio.isAttributeOf, 'Attribute': rdf.type, 'Definition' : skos.definition, 'Value' : sio.hasValue, 'wasDerivedFrom': prov.wasDerivedFrom, 'Label': rdfs.label, 'inRelationTo': sio.inRelationTo, 'Role': sio.hasRole, 'Start' : sio.hasStartTime, 'End' : sio.hasEndTime, 'Time': sio.existsAt, 'Entity': rdf.type, 'Unit': sio.hasUnit, 'wasGeneratedBy': prov.wasGeneratedBy}
if 'properties' in config['Source Files'] :
properties_fn = config['Source Files']['properties']
try :
properties_file = pd.read_csv(properties_fn, dtype=object)
except Exception as e :
print("Warning: The specified Properties file does not exist or is unreadable: " + str(e))
return properties_tuple
for row in properties_file.itertuples() :
if(hasattr(row,"Property") and pd.notnull(row.Property)):
properties_tuple[row.Column]=row.Property
return properties_tuple
def processTimeline(timeline_fn):
timeline_tuple = {}
if timeline_fn is not None :
try :
timeline_file = pd.read_csv(timeline_fn, dtype=object)
try :
inner_tuple_list = []
row_num=0
for row in timeline_file.itertuples():
if (pd.notnull(row.Name) and row.Name not in timeline_tuple) :
inner_tuple_list=[]
inner_tuple = {}
inner_tuple["Type"]=row.Type
if(hasattr(row,"Label") and pd.notnull(row.Label)):
inner_tuple["Label"]=row.Label
if(pd.notnull(row.Start)) :
inner_tuple["Start"]=row.Start
if( | pd.notnull(row.End) | pandas.notnull |
import pandas as pd
import pathlib
from scripts.python.routines.mvals import logit2
import numpy as np
path_global = f"E:/YandexDisk/Work/pydnameth/datasets"
folder_name = f"GPL13534_Blood_ICD10-V"
path = f"{path_global}/meta/tasks/GPL13534_Blood_ICD10-V"
pathlib.Path(f"{path}/R/one_by_one").mkdir(parents=True, exist_ok=True)
betas = | pd.read_pickle(f"{path}/train_val/betas.pkl") | pandas.read_pickle |
import math
import warnings
import numpy as np
import pandas as pd
import scipy.signal
import matplotlib.pyplot as plt
from typing import Optional, Union, List
from tqdm import tqdm
from signalanalysis.signalanalysis import general
from signalanalysis import signalplot
from signalanalysis import tools
class Egm(general.Signal):
"""Base class for EGM data, inheriting from :class:`signalanalysis.signalanalysis.general.Signal`
See Also
--------
:py:class:`signalanalysis.signalanalysis.general.Signal`
Methods
-------
read(folder)
Extract data from unipolar and bipolar DxL files
get_n_beats()
Supersedes generalised method to calculate n_beats
get_at
Calculates the activation time of the EGM
"""
def __init__(self,
data_location_uni: str,
data_location_bi: str = None,
**kwargs):
"""Sub-method for __init___
Will initialise a EGM signal class
TODO: Fix the self.data reference problem (see
https://stackoverflow.com/questions/6057130/python-deleting-a-class-attribute-in-a-subclass)
See Also
--------
:py:meth:`signalanalysis.signalanalysis.general.Signal.__init__ : Base __init__ method
:py:meth:`signalanalysis.signalanalysis.general.Signal.apply_filter` : Filtering method
:py:meth:`signalanalysis.signalanalysis.general.Signal.get_n_beats` : Beat calculation method
Notes
-----
This used to break the `Liskov substitution principle
<https://en.wikipedia.org/wiki/Liskov_substitution_principle>`_, removing the single `data` attribute to be
replaced by `data_uni` and `data_bi`, but now instead (aims to) just point the `.data` attribute to the
`.data_uni` attribute
"""
super(Egm, self).__init__(**kwargs)
self.t_peaks = pd.DataFrame(dtype=float)
self.n_beats = pd.Series(dtype=int)
# delattr(self, 'data')
self.data_uni = pd.DataFrame(dtype=float)
self.data_bi = pd.DataFrame(dtype=float)
self.beats_uni = dict()
self.beats = self.beats_uni
self.beats_bi = dict()
self.at = pd.DataFrame(dtype=float)
self.rt = pd.DataFrame(dtype=float)
self.ari = pd.DataFrame(dtype=float)
self.dvdt = pd.DataFrame(dtype=float)
self.qrs_start = pd.DataFrame(dtype=float)
self.qrs_end = pd.DataFrame(dtype=float)
self.qrs_duration = pd.DataFrame(dtype=float)
self.read(data_location_uni, data_location_bi, **kwargs)
if self.filter is not None:
self.apply_filter(**kwargs)
self.data = self.data_uni
# self.get_beats(**kwargs)
def read(self,
data_location_uni: str,
data_location_bi: Optional[str] = None,
drop_empty_rows: bool = True,
**kwargs):
""" Read the DxL data for unipolar and bipolar data for EGMs
TODO: Add functionality to read directly from folders, rather than .csv from Matlab
Parameters
----------
data_location_uni : str
Location of unipolar data. Currently only coded to deal with a saved .csv file
data_location_bi : str, optional
Location of bipolar data. Currently only coded to deal with a saved .csv file. Doesn't need to be passed,
default=None
drop_empty_rows : bool, optional
Whether to drop empty data rows from the data, default=True
See Also
--------
:py:meth:`signalanalysis.signalanalysis.egm.Egm.read_from_csv` : Method to read data from Matlab csv
"""
if data_location_uni.endswith('.csv'):
if data_location_bi is not None:
assert data_location_bi.endswith('.csv')
self.read_from_csv(data_location_uni, data_location_bi, **kwargs)
else:
raise IOError("Not coded for this type of input")
if drop_empty_rows:
# PyCharm highlights an error below (bool doesn't have a .all() method), but I'll be damned if I can
# figure out how to fix it - the below option replaces *all* 0.00 values, so will put NaN in an otherwise
# normal trace where it happens to reach 0.00, which is not what we want.
# self.data_uni = (self.data_uni.where(self.data_uni != 0, axis=0)).dropna(axis=1, how='all')
self.data_uni = self.data_uni.loc[:, ~(self.data_uni == 0).all(axis=0)]
if not self.data_bi.empty:
self.data_bi = self.data_bi.loc[:, ~(self.data_bi == 0).all(axis=0)]
assert self.data_uni.shape == self.data_bi.shape, "Error in dropping rows"
return None
def read_from_csv(self,
data_location_uni: str,
data_location_bi: Optional[str],
frequency: float):
""" Read EGM data that has been saved from Matlab
Parameters
----------
data_location_uni : str
Name of the .csv file containing the unipolar data
data_location_bi : str, optional
Name of the .csv file containing the bipolar data
frequency : float
The frequency of the data recording in Hz
Notes
-----
It is not technically required to pass the bipolar data, but it is presented here as a required keyword to
preserve the usage of calling as `read_from_csv(unipolar, bipolar, frequency)`, rather than breaking the data
files arguments up or requiring keywords.
The .csv file should be saved with column representing an individual EGM trace, and each row representing a
single instance in time, i.e.
.. code-block::
egm1(t1), egm2(t1), egm3(t1), ...
egm1(t2), egm2(t2), egm3(t2), ...
...
egm1(tn), egm2(tn), egm3(tn)
Historically, `frequency` has been set to 2034.5 Hz for the importprecision data, an example of which is
can be accessed via ``signalanalysis.data.datafiles.EGM_UNIPOLAR`` and ``signalanalysis.data.datafiles.EGM_BIPOLAR``.
"""
self.data_uni = pd.read_csv(data_location_uni, header=None)
interval = (1 / frequency)*1000
end_val = self.data_uni.shape[0] * interval
t = np.arange(0, end_val, interval)
self.data_uni.set_index(t, inplace=True)
if data_location_bi is not None:
self.data_bi = pd.read_csv(data_location_bi, header=None)
self.data_bi.set_index(t, inplace=True)
self.data_source = [data_location_uni, data_location_bi]
else:
self.data_bi = pd.DataFrame()
self.data_source = data_location_uni
return None
def get_peaks(self,
threshold: float = 0.33,
min_separation: float = 200,
plot: bool = False,
**kwargs):
""" Supermethod for get_peaks for EGM data, using the squared bipolar signal rather than RMS data
See also
--------
:py:meth:`signalanalysis.signalanalysis.egm.Egm.plot_signal` : Method to plot the calculated AT
"""
if self.data_bi.empty:
# super(Egm, self).get_peaks()
egm_bi_square = np.abs(self.data_uni)
else:
egm_bi_square = np.square(self.data_bi)
i_separation = np.where(self.data_uni.index > min_separation)[0][0]
self.n_beats = pd.Series(dtype=int, index=self.data_uni.columns)
self.t_peaks = pd.DataFrame(dtype=float, columns=self.data_uni.columns)
self.n_beats_threshold = threshold
for i_signal in egm_bi_square:
i_peaks, _ = scipy.signal.find_peaks(egm_bi_square.loc[:, i_signal],
height=threshold*egm_bi_square.loc[:, i_signal].max(),
distance=i_separation)
self.n_beats[i_signal] = len(i_peaks)
# Pad the peaks data or t_peaks dataframe with NaN as appropriate
if len(i_peaks) == self.t_peaks.shape[0]:
self.t_peaks[i_signal] = self.data_uni.index[i_peaks]
elif len(i_peaks) < self.t_peaks.shape[0]:
self.t_peaks[i_signal] = np.pad(self.data_uni.index[i_peaks],
(0, self.t_peaks.shape[0]-len(i_peaks)),
constant_values=float("nan"))
elif len(i_peaks) > self.t_peaks.shape[0]:
self.t_peaks = self.t_peaks.reindex(range(len(i_peaks)), fill_value=float("nan"))
self.t_peaks[i_signal] = self.data_uni.index[i_peaks]
if plot:
_ = signalplot.egm.plot_signal(self, plot_peaks=True, plot_bipolar_square=True, **kwargs)
return None
def get_beats(self,
reset_index: bool = True,
offset_start: Optional[float] = None,
offset_end: Optional[float] = None,
plot: bool = False,
**kwargs):
""" Detects beats in individual EGM signals
TODO: Replace this with method based on finding AT and RT, then adding buffer round those values
Supermethod for EGM beat detection, due to the fact that EGM beats are detected on a per signal basis
rather than a universal basis (RMS method)
See also
--------
:py:meth:`signalanalysis.signalanalysis.general.Signal.get_beats` : Base method
"""
if self.t_peaks.empty:
self.get_peaks(**kwargs)
# we'll store these values in data frames later on
beat_start_values = np.full_like(self.t_peaks, fill_value=np.NaN)
beat_end_values = np.full_like(self.t_peaks, fill_value=np.NaN)
self.beats_uni = dict.fromkeys(self.data_uni.columns)
self.beats_bi = dict.fromkeys(self.data_uni.columns)
all_bcls = np.diff(self.t_peaks, axis=0).T
for key, bcls in zip(self.data_uni, all_bcls):
# If only one beat is detected, can end here
n_beats = self.n_beats[key]
if n_beats == 1:
self.beats_uni[key] = [self.data_uni.loc[:, key]]
self.beats_bi[key] = [self.data_bi.loc[:, key]]
continue
# Calculate series of cycle length values, before then using this to estimate the start and end times of
# each beat. The offset from the previous peak will be assumed at 0.4*BCL, while the offset from the
# following peak will be 0.1*BCL (both with a minimum value of 30ms)
if offset_start is None:
offset_start_list = [max(0.6 * bcl, 30) for bcl in bcls[:n_beats-1]]
else:
offset_start_list = [offset_start] * (self.n_beats[key] - 1)
if offset_end is None:
offset_end_list = [max(0.1 * bcl, 30) for bcl in bcls[:n_beats-1]]
else:
offset_end_list = [offset_end] * (self.n_beats[key] - 1)
beat_start = [self.data_uni.index[0]]
beat_start.extend(self.t_peaks[key][:n_beats-1].values + offset_start_list)
beat_end = []
beat_end.extend(self.t_peaks[key][1:n_beats].values - offset_end_list)
beat_end.append(self.data_uni.index[-1])
# we'll store these values in data frames later on
column_index = self.t_peaks.columns.get_loc(key)
beat_start_values[:n_beats, column_index] = beat_start
beat_end_values[:n_beats, column_index] = beat_end
signal_beats_uni = np.empty(n_beats, dtype=object)
signal_beats_bi = np.empty(n_beats, dtype=object)
for beat_index, (t_s, t_p, t_e) in enumerate(zip(beat_start, self.t_peaks[key], beat_end)):
if not (t_s < t_p < t_e):
raise ValueError("Error in windowing process - a peak is outside of the window for EGM ", key)
signal_beats_uni[beat_index] = self.data_uni.loc[t_s:t_e, :]
signal_beats_bi[beat_index] = self.data_bi.loc[t_s:t_e, :]
if not reset_index:
continue
zeroed_index = signal_beats_uni[beat_index].index - signal_beats_uni[beat_index].index[0]
signal_beats_uni[beat_index].set_index(zeroed_index, inplace=True)
signal_beats_bi[beat_index].set_index(zeroed_index, inplace=True)
self.beat_index_reset = reset_index
self.beats_uni[key] = signal_beats_uni
self.beats_bi[key] = signal_beats_bi
self.beat_start = pd.DataFrame(
data=beat_start_values,
index=self.t_peaks.index,
columns=self.t_peaks.columns,
dtype=float,
)
self.beat_end = pd.DataFrame(
data=beat_end_values,
index=self.t_peaks.index,
columns=self.t_peaks.columns,
dtype=float,
)
if plot:
_ = self.plot_beats(offset_end=offset_end, **kwargs)
def plot_beats(self,
i_plot: Optional[int] = None,
**kwargs):
"""
..deprecated::
Need to move this to signalanalysis.signalplot.egm (if this even works!)
"""
# Calculate beats (if not done already)
if self.beats_uni is None:
self.get_beats(offset_end=None, plot=False, **kwargs)
# Pick a random signal to plot as an example trace (making sure to not pick a 'dead' trace)
if i_plot is None:
weights = (self.n_beats.values > 0).astype(int)
i_plot = self.n_beats.sample(weights=weights).index[0]
elif self.n_beats[i_plot] == 0:
raise IOError("No beats detected in specified trace")
n_beats = n_beats = self.n_beats[i_plot]
t_peaks = self.t_peaks[i_plot]
beat_start = self.beat_start[i_plot]
beat_end = self.beat_end[i_plot]
ax_labels = ['Unipolar', 'Bipolar']
egm_data = [self.data_uni, self.data_bi]
colours = tools.plotting.get_plot_colours(n_beats)
fig, axes = plt.subplots(2, 1)
fig.suptitle('Trace {}'.format(i_plot))
for index, (ax, data) in enumerate(zip(axes, egm_data)):
plt.sca(ax)
plt.plot(data.loc[:, i_plot], color='C0')
plt.scatter(
t_peaks[:n_beats],
data.loc[:, i_plot][t_peaks[:n_beats]],
marker='o',
edgecolor='tab:orange',
facecolor='none',
linewidths=2,
)
plt.ylabel(ax_labels[index])
max_height = np.max(data.loc[:, i_plot])
height_shift = (np.max(data.loc[:, i_plot]) - np.min(data.loc[:, i_plot])) * 0.1
height_val = [max_height, max_height - height_shift] * math.ceil(n_beats / 2)
for beat_index, (t_s, t_e) in enumerate(zip(beat_start[:n_beats], beat_end[:n_beats])):
plt.axvline(t_s, color=colours[beat_index])
plt.axvline(t_e, color=colours[beat_index])
plt.annotate(text='{}'.format(beat_index+1), xy=(t_s, height_val[beat_index]), xytext=(t_e, height_val[beat_index]),
arrowprops=dict(arrowstyle='<->', linewidth=3))
return fig, ax
def get_at(self,
at_window: float = 30,
unipolar_delay: float = 50,
plot: bool = False,
**kwargs):
""" Calculates the activation time for a given beat of EGM data
Will calculate the activation times for an EGM signal, based on finding the peaks in the squared bipolar
trace, then finding the maximum downslope in the unipolar signal within a specified window of time around
those peaks. Note that, if bipolar data are not present, then the squared unipolar signal will be used,
which will invariably find the pacing artefact. As such, when unipolar peaks are used, a 'delay' will be
applied to the window to avoid the pacing artefact.
Parameters
----------
at_window : float, optional
Time in milliseconds, around which the activation time will be searched for round the detected peaks,
i.e. the EGM trace will be searched in the window t_peak +/- at_window. Default=30ms
unipolar_delay : float, optional
Time in milliseconds to delay the search window after the peak time, if only unipolar data are being
used, to avoid getting confused with the far-field pacing artefact. Will thus have the search window
adapted to (t_peak+unipolar_delay) +/- at_window. Default=50ms
plot : bool, optional
Whether to plot a random signal example of the ATs found, default=False
See also
--------
:py:meth:`signalanalysis.signalanalysis.egm.Egm.get_peaks` : Method to calculate peaks
:py:meth:`signalanalysis.signalanalysis.egm.Egm.plot_signal` : Method to plot the signal
"""
if self.t_peaks.empty:
self.get_peaks()
egm_uni_grad_full = pd.DataFrame(np.gradient(self.data_uni, axis=0),
index=self.data_uni.index,
columns=self.data_uni.columns)
# Calculate and adjust the start and end point for window searches
if not self.data_bi.empty:
unipolar_delay = 0
window_start = self.return_to_index(self.t_peaks.sub(at_window).add(unipolar_delay))
window_end = self.return_to_index(self.t_peaks.add(at_window).add(unipolar_delay))
self.at = self.t_peaks.copy()
# Current brute force method
from tqdm import tqdm
for key in tqdm(window_start, desc='Finding AT...', total=len(window_start.columns)):
for i_row, _ in window_start[key].iteritems():
t_s = window_start.loc[i_row, key]
if pd.isna(t_s):
continue
t_e = window_end.loc[i_row, key]
self.at.loc[i_row, key] = egm_uni_grad_full.loc[t_s:t_e, key].idxmin()
self.dvdt.loc[i_row, key] = egm_uni_grad_full.loc[t_s:t_e, key].min()
if plot:
_ = signalplot.egm.plot_signal(self, plot_at=True, **kwargs)
return None
def get_rt(self,
lower_window_limit: float = 140,
plot: bool = False,
**kwargs):
""" Calculate the repolarisation time
Calculates the repolarisation time of an action potential from the EGM, based on the Wyatt method of the
maximum upslope of the T-wave
TODO: try to improve on the current brute force method used to find the point of RT
Parameters
----------
lower_window_limit : float, optional
Minimum time after the AT to have passed before repolarisation can potentially happen, default=150ms
plot : bool, optional
Whether to plot a random signal example of the ATs found, default=False
Returns
-------
self.rt : pd.DataFrame
Repolarisation times for each signal in the trace
self.ari : pd.DataFrame
Activation repolarisation intervals for each AT/RT pair
References
----------
.. [1] <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>,
<NAME>, <NAME>, <NAME>, "Autonomic modulation in patients with heart failure increases
beat-to-beat variability of ventricular action potential duration. Frontiers in Physiology, 8(MAY 2017).
https://doi.org/10.3389/fphys.2017.00328
"""
# Estimate BCL, then calculate the upper and lower bounds within which to search for the repolarisation time
if self.at.empty:
self.get_at(**kwargs)
bcl = general.get_bcl(self.at)
# INITIALISE WINDOWS WITHIN WHICH TO SEARCH FOR RT
window_start = (bcl.mul(0.75)).sub(125) # Equivalent to 0.75*bcl-125
window_start[window_start < lower_window_limit] = lower_window_limit
window_start = self.at+window_start
window_end = (bcl.mul(0.9)).sub(50) # Equivalent to 0.9*bcl-50
window_end = self.at+window_end
window_end[window_end-window_start < 0.1] = window_start+0.1
# If the end of the search window is within 20ms of the next AT/end of the recording, shorten the end of the
# window accordingly
# Don't bother looking for RT if the start of the search window is within 40ms of the following AT/the end of
# the recording.
def window_max_generator(buffer):
window_max = self.at - buffer
window_max.set_index(window_max.index - 1, inplace=True)
window_max.drop(-1, inplace=True)
window_max = window_max.append(pd.DataFrame(self.data_uni.index[-1] - buffer,
columns=window_max.columns,
index=[window_max.index[-1] + 1]))
window_max[window_max > self.data_uni.index[-1] - buffer] = \
self.data_uni.index[-1] - buffer
window_max.fillna(axis=0, method='bfill', inplace=True)
return window_max
window_start_max = window_max_generator(40)
window_start[window_start > window_start_max] = float("nan")
window_start = self.return_to_index(window_start)
window_end_max = window_max_generator(20)
window_end[window_end > window_end_max] = window_end_max
window_end = self.return_to_index(window_end)
# Brute force method!
egm_uni_grad = pd.DataFrame(np.gradient(self.data_uni, axis=0),
index=self.data_uni.index,
columns=self.data_uni.columns)
self.rt = pd.DataFrame(index=self.at.index, columns=self.at.columns)
self.ari = pd.DataFrame(index=self.at.index, columns=self.at.columns)
for key in tqdm(window_start, desc='Finding RT...', total=len(window_start.columns)):
for i_row, _ in enumerate(window_start[key]):
# FIND T-WAVE PEAK
# Look for the peak of the unipolar EGM within the search window. If the maximum (+/- 0.03mV) is at the
# start/end of the window, shorten the window and check again to try and ensure that the peak
# represents the T-wave peak rather than the repolarisation/depolarisation preceding/following the
# T-wave.
window_error = False
negative_t_wave = False
t_start = window_start.loc[i_row, key]
t_end = window_end.loc[i_row, key]
if pd.isna(t_start) or pd.isna(t_end):
continue
i_ts = np.where(self.data_uni.index.values == t_start)[0]
i_te = np.where(self.data_uni.index.values == t_end)[0]
uni_start = self.data_uni.loc[t_start, key]
uni_end = self.data_uni.loc[t_end, key]
uni_peak = self.data_uni.loc[t_start:t_end, key].max()
uni_start_diff = abs(uni_start-uni_peak)
uni_end_diff = abs(uni_end-uni_peak)
while uni_start_diff <= 0.03 or uni_end_diff <= 0.03:
while uni_start_diff <= 0.03:
i_ts = i_ts+1
try:
t_start = self.data_uni.index[i_ts][0]
except IndexError:
pass
if t_start >= t_end:
window_error = True
break
uni_start = self.data_uni.loc[t_start, key]
uni_peak = self.data_uni.loc[t_start:t_end, key].max()
uni_start_diff = abs(uni_start-uni_peak)
if window_error:
break
while uni_end_diff <= 0.03:
i_te = i_te-1
try:
t_end = self.data_uni.index[i_te][0]
except IndexError:
pass
if t_start >= t_end:
window_error = True
break
uni_end = self.data_uni.loc[t_end, key]
uni_peak = self.data_uni.loc[t_start:t_end, key].max()
uni_end_diff = abs(uni_end-uni_peak)
# If it is impossible to narrow the search window as above and find a positive peak for the T-wave,
# set the window to the original values and assume that the T-wave is negative
if window_error or not (egm_uni_grad.loc[t_start:t_end, key] > 0).any():
t_start = window_start.loc[i_row, key]
t_end = window_start.loc[i_row, key]
t_peak = self.data_uni.loc[t_start:t_end, key].idxmin()
negative_t_wave = True
else:
t_peak = self.data_uni.loc[t_start:t_end, key].idxmax()
assert t_start <= t_peak <= t_end, "Problem setting window values"
# FIND REPOLARISATION TIME
max_grad = -100
t_max_grad = -1
window_data = egm_uni_grad.loc[t_start:t_end, key]
t_index_in_uni_data = np.searchsorted(self.data_uni.index.values, window_data.index.values)
for (t_window, uni_val), i_tm in zip(window_data.iteritems(), t_index_in_uni_data):
# Look for maximum gradient in the search window thus far
if uni_val > max_grad:
max_grad = uni_val
t_max_grad = t_window
# Perform check to see if we've exited the current T-wave (if we're after the total max peak
# (apex) and have negative gradient)
if negative_t_wave:
self.rt.loc[i_row, key] = t_max_grad
self.ari.loc[i_row, key] = t_max_grad - self.at.loc[i_row, key]
else:
t1 = self.data_uni.index[i_tm-1]
t2 = self.data_uni.index[i_tm+2] # Adding 2 to ensure that the limit is taken at +1, not i_tm
if (t_window > t_peak) and (window_data.loc[t1:t2] < 0).all():
self.rt.loc[i_row, key] = t_max_grad
self.ari.loc[i_row, key] = t_max_grad - self.at.loc[i_row, key]
break
if plot:
_ = signalplot.egm.plot_signal(self, plot_rt=True, **kwargs)
return None
def get_ari(self,
plot: bool = False,
**kwargs):
"""Dummy function to calculate ARI
TODO: check that `plot` keyword is correctly over-ridden (if that is possible)
ARI is calculated as part of self.get_rt, so this module serves just as useful syntax.
See also
--------
:py:meth:`signalanalysis.signalanalysis.egm.Egm.get_rt` : Actual method called
"""
if self.ari.empty:
self.get_rt(plot=False, **kwargs)
if plot:
signalplot.egm.plot_signal(plot_at=True, plot_rt=True, **kwargs)
return None
def get_qrsd(self,
lower_window: float = 30,
upper_window: float = 60,
threshold: float = 0.1,
plot: bool = True,
**kwargs):
"""Calculates the QRS duration for EGM data
TODO: See if this can be improved beyond the current brute force method
The start and end of the QRS complex is calculated as the duration for which the energy of the bipolar signal
(defined as the bipolar signal squared) exceeds a threshold value. The 'window' over which to search for this
complex is defined from the detected activation times, plus/minus specified values (`lower_window` and
`upper_window`). Note that, due to the calculation method, this cannot be calculated for instances where no
bipolar data are available.
Parameters
----------
lower_window, upper_window : float, optional
Window before/after AT to search for QRS start/end, respectively, given in milliseconds, default=30/60ms
threshold : float, optional
Fractional threshold of maximum energy used to define the start and end of the QRS complex, default=0.1
plot : bool, optional
Whether or not to plot an example trace
Returns
-------
self.qrs_duration : pd.DataFrame
QRS durations for each signal
See also
--------
:py:meth:`signalanalysis.signalanalysis.egm.Egm.get_at` : Method used to calculate AT, that uses this method implicitly
:py:meth:`signalanalysis.signalplot.egm.plot_signal` : Plotting function, with options that can be passed in **kwargs
"""
if self.data_bi.empty:
raise IOError('Cannot calculate QRSd for unipolar only data')
# Sanitise inputs and make sure they make sense
if lower_window < 0.5:
warnings.warn('Assuming that lWindow has been entered in seconds rather than milliseconds: correcting...')
lower_window = lower_window * 1000
if upper_window < 0.5:
warnings.warn('Assuming that uWindow has been entered in seconds rather than milliseconds: correcting...')
upper_window = upper_window * 1000
assert 0.0 < threshold < 1.0, "threshold must be set between 0 and 1"
if self.at.empty:
self.get_at(**kwargs)
window_start = self.return_to_index(self.at.sub(lower_window))
window_end = self.return_to_index(self.at.add(upper_window))
for key in tqdm(self.at, desc='Finding QRSd...', total=len(self.at.columns)):
for i_row, _ in enumerate(self.at[key]):
# Only continue if the window values aren't NaN
if pd.isna(window_start.loc[i_row, key]) or | pd.isna(window_end.loc[i_row, key]) | pandas.isna |
import streamlit as st
import pandas as pd, seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
import joblib
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import scipy.sparse as sp
import pytesseract
pytesseract.pytesseract.tesseract_cmd = r"D:\Programming Softwares\New folder\tesseract.exe"
import tensorflow as tf
import re
import nltk
from nltk.corpus import stopwords
# nltk.download('stopwords')
from PIL import Image
import plotly.express as px
from plotly.subplots import make_subplots
import plotly.graph_objects as go
import plotly
from plotly import tools
stopwords_nltk = stopwords.words("english")
new_stopwords = ['filtered','including','every','actual','equivalent', 'less','contains','actual',"concentrate","100","artificial","coloring","simple","medium","chain","flavorings","flavor",""]
stopwords_nltk.extend(new_stopwords)
####################################################################################################
#############-----------------------------LOAD DATA----------------------------------###############
####################################################################################################
@st.cache(show_spinner=False)
def load_data():
# rec_path = r"D:\CoderSchool_ML30\FINAL PROJECT\Data\recommendation_df.csv"
rec_path = r"D:\CoderSchool_ML30\FINAL PROJECT\Data\rec_df.csv"
tfidf_df = pd.read_csv(rec_path)
tfidf_df = tfidf_df.drop(columns=["New Ingredients"])
df = pd.read_csv(r"D:\CoderSchool_ML30\FINAL PROJECT\Data\eda.csv")
veg_df = pd.read_csv(r"D:\CoderSchool_ML30\FINAL PROJECT\Data\veg_df.csv")
veg_df = veg_df[~veg_df.Name.duplicated()]
additives_count = pd.read_csv(r"D:\CoderSchool_ML30\FINAL PROJECT\Data\additives_count.csv")
additives_count = additives_count.sort_values("Count")
add_df = | pd.read_csv(r"D:\CoderSchool_ML30\FINAL PROJECT\Data\OCR_additives.csv") | pandas.read_csv |
"""AWS Glue Catalog Module."""
# pylint: disable=redefined-outer-name
import itertools
import logging
import re
import unicodedata
from typing import Any, Dict, Iterator, List, Optional, Tuple
from urllib.parse import quote_plus
import boto3 # type: ignore
import pandas as pd # type: ignore
import sqlalchemy # type: ignore
from awswrangler import _data_types, _utils, exceptions
_logger: logging.Logger = logging.getLogger(__name__)
def delete_table_if_exists(database: str, table: str, boto3_session: Optional[boto3.Session] = None) -> bool:
"""Delete Glue table if exists.
Parameters
----------
database : str
Database name.
table : str
Table name.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
bool
True if deleted, otherwise False.
Examples
--------
>>> import awswrangler as wr
>>> wr.catalog.delete_table_if_exists(database='default', name='my_table') # deleted
True
>>> wr.catalog.delete_table_if_exists(database='default', name='my_table') # Nothing to be deleted
False
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
try:
client_glue.delete_table(DatabaseName=database, Name=table)
return True
except client_glue.exceptions.EntityNotFoundException:
return False
def does_table_exist(database: str, table: str, boto3_session: Optional[boto3.Session] = None):
"""Check if the table exists.
Parameters
----------
database : str
Database name.
table : str
Table name.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
bool
True if exists, otherwise False.
Examples
--------
>>> import awswrangler as wr
>>> wr.catalog.does_table_exist(database='default', name='my_table')
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
try:
client_glue.get_table(DatabaseName=database, Name=table)
return True
except client_glue.exceptions.EntityNotFoundException:
return False
def create_parquet_table(
database: str,
table: str,
path: str,
columns_types: Dict[str, str],
partitions_types: Optional[Dict[str, str]] = None,
compression: Optional[str] = None,
description: Optional[str] = None,
parameters: Optional[Dict[str, str]] = None,
columns_comments: Optional[Dict[str, str]] = None,
mode: str = "overwrite",
boto3_session: Optional[boto3.Session] = None,
) -> None:
"""Create a Parquet Table (Metadata Only) in the AWS Glue Catalog.
'https://docs.aws.amazon.com/athena/latest/ug/data-types.html'
Parameters
----------
database : str
Database name.
table : str
Table name.
path : str
Amazon S3 path (e.g. s3://bucket/prefix/).
columns_types: Dict[str, str]
Dictionary with keys as column names and vales as data types (e.g. {'col0': 'bigint', 'col1': 'double'}).
partitions_types: Dict[str, str], optional
Dictionary with keys as partition names and values as data types (e.g. {'col2': 'date'}).
compression: str, optional
Compression style (``None``, ``snappy``, ``gzip``, etc).
description: str, optional
Table description
parameters: Dict[str, str], optional
Key/value pairs to tag the table.
columns_comments: Dict[str, str], optional
Columns names and the related comments (e.g. {'col0': 'Column 0.', 'col1': 'Column 1.', 'col2': 'Partition.'}).
mode: str
'overwrite' to recreate any possible existing table or 'append' to keep any possible existing table.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
None
None.
Examples
--------
>>> import awswrangler as wr
>>> wr.catalog.create_parquet_table(
... database='default',
... table='my_table',
... path='s3://bucket/prefix/',
... columns_types={'col0': 'bigint', 'col1': 'double'},
... partitions_types={'col2': 'date'},
... compression='snappy',
... description='My own table!',
... parameters={'source': 'postgresql'},
... columns_comments={'col0': 'Column 0.', 'col1': 'Column 1.', 'col2': 'Partition.'}
... )
"""
table = sanitize_table_name(table=table)
partitions_types = {} if partitions_types is None else partitions_types
table_input: Dict[str, Any] = _parquet_table_definition(
table=table, path=path, columns_types=columns_types, partitions_types=partitions_types, compression=compression
)
_create_table(
database=database,
table=table,
description=description,
parameters=parameters,
columns_comments=columns_comments,
mode=mode,
boto3_session=boto3_session,
table_input=table_input,
)
def _parquet_table_definition(
table: str, path: str, columns_types: Dict[str, str], partitions_types: Dict[str, str], compression: Optional[str]
) -> Dict[str, Any]:
compressed: bool = compression is not None
return {
"Name": table,
"PartitionKeys": [{"Name": cname, "Type": dtype} for cname, dtype in partitions_types.items()],
"TableType": "EXTERNAL_TABLE",
"Parameters": {"classification": "parquet", "compressionType": str(compression).lower(), "typeOfData": "file"},
"StorageDescriptor": {
"Columns": [{"Name": cname, "Type": dtype} for cname, dtype in columns_types.items()],
"Location": path,
"InputFormat": "org.apache.hadoop.hive.ql.io.parquet.MapredParquetInputFormat",
"OutputFormat": "org.apache.hadoop.hive.ql.io.parquet.MapredParquetOutputFormat",
"Compressed": compressed,
"NumberOfBuckets": -1,
"SerdeInfo": {
"SerializationLibrary": "org.apache.hadoop.hive.ql.io.parquet.serde.ParquetHiveSerDe",
"Parameters": {"serialization.format": "1"},
},
"StoredAsSubDirectories": False,
"SortColumns": [],
"Parameters": {
"CrawlerSchemaDeserializerVersion": "1.0",
"classification": "parquet",
"compressionType": str(compression).lower(),
"typeOfData": "file",
},
},
}
def add_parquet_partitions(
database: str,
table: str,
partitions_values: Dict[str, List[str]],
compression: Optional[str] = None,
boto3_session: Optional[boto3.Session] = None,
) -> None:
"""Add partitions (metadata) to a Parquet Table in the AWS Glue Catalog.
Parameters
----------
database : str
Database name.
table : str
Table name.
partitions_values: Dict[str, List[str]]
Dictionary with keys as S3 path locations and values as a list of partitions values as str
(e.g. {'s3://bucket/prefix/y=2020/m=10/': ['2020', '10']}).
compression: str, optional
Compression style (``None``, ``snappy``, ``gzip``, etc).
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
None
None.
Examples
--------
>>> import awswrangler as wr
>>> wr.catalog.add_parquet_partitions(
... database='default',
... table='my_table',
... partitions_values={
... 's3://bucket/prefix/y=2020/m=10/': ['2020', '10'],
... 's3://bucket/prefix/y=2020/m=11/': ['2020', '11'],
... 's3://bucket/prefix/y=2020/m=12/': ['2020', '12']
... }
... )
"""
inputs: List[Dict[str, Any]] = [
_parquet_partition_definition(location=k, values=v, compression=compression)
for k, v in partitions_values.items()
]
_add_partitions(database=database, table=table, boto3_session=boto3_session, inputs=inputs)
def _parquet_partition_definition(location: str, values: List[str], compression: Optional[str]) -> Dict[str, Any]:
compressed: bool = compression is not None
return {
"StorageDescriptor": {
"InputFormat": "org.apache.hadoop.hive.ql.io.parquet.MapredParquetInputFormat",
"OutputFormat": "org.apache.hadoop.hive.ql.io.parquet.MapredParquetOutputFormat",
"Location": location,
"Compressed": compressed,
"SerdeInfo": {
"Parameters": {"serialization.format": "1"},
"SerializationLibrary": "org.apache.hadoop.hive.ql.io.parquet.serde.ParquetHiveSerDe",
},
"StoredAsSubDirectories": False,
},
"Values": values,
}
def get_table_types(database: str, table: str, boto3_session: Optional[boto3.Session] = None) -> Dict[str, str]:
"""Get all columns and types from a table.
Parameters
----------
database : str
Database name.
table : str
Table name.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
Dict[str, str]
A dictionary as {'col name': 'col data type'}.
Examples
--------
>>> import awswrangler as wr
>>> wr.catalog.get_table_types(database='default', name='my_table')
{'col0': 'int', 'col1': double}
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
response: Dict[str, Any] = client_glue.get_table(DatabaseName=database, Name=table)
dtypes: Dict[str, str] = {}
for col in response["Table"]["StorageDescriptor"]["Columns"]:
dtypes[col["Name"]] = col["Type"]
for par in response["Table"]["PartitionKeys"]:
dtypes[par["Name"]] = par["Type"]
return dtypes
def get_databases(
catalog_id: Optional[str] = None, boto3_session: Optional[boto3.Session] = None
) -> Iterator[Dict[str, Any]]:
"""Get an iterator of databases.
Parameters
----------
catalog_id : str, optional
The ID of the Data Catalog from which to retrieve Databases.
If none is provided, the AWS account ID is used by default.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
Iterator[Dict[str, Any]]
Iterator of Databases.
Examples
--------
>>> import awswrangler as wr
>>> dbs = wr.catalog.get_databases()
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
paginator = client_glue.get_paginator("get_databases")
if catalog_id is None:
response_iterator: Iterator = paginator.paginate()
else:
response_iterator = paginator.paginate(CatalogId=catalog_id)
for page in response_iterator:
for db in page["DatabaseList"]:
yield db
def databases(
limit: int = 100, catalog_id: Optional[str] = None, boto3_session: Optional[boto3.Session] = None
) -> pd.DataFrame:
"""Get a Pandas DataFrame with all listed databases.
Parameters
----------
limit : int, optional
Max number of tables to be returned.
catalog_id : str, optional
The ID of the Data Catalog from which to retrieve Databases.
If none is provided, the AWS account ID is used by default.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
pandas.DataFrame
Pandas DataFrame filled by formatted infos.
Examples
--------
>>> import awswrangler as wr
>>> df_dbs = wr.catalog.databases()
"""
database_iter: Iterator[Dict[str, Any]] = get_databases(catalog_id=catalog_id, boto3_session=boto3_session)
dbs = itertools.islice(database_iter, limit)
df_dict: Dict[str, List] = {"Database": [], "Description": []}
for db in dbs:
df_dict["Database"].append(db["Name"])
if "Description" in db:
df_dict["Description"].append(db["Description"])
else: # pragma: no cover
df_dict["Description"].append("")
return pd.DataFrame(data=df_dict)
def get_tables(
catalog_id: Optional[str] = None,
database: Optional[str] = None,
name_contains: Optional[str] = None,
name_prefix: Optional[str] = None,
name_suffix: Optional[str] = None,
boto3_session: Optional[boto3.Session] = None,
) -> Iterator[Dict[str, Any]]:
"""Get an iterator of tables.
Parameters
----------
catalog_id : str, optional
The ID of the Data Catalog from which to retrieve Databases.
If none is provided, the AWS account ID is used by default.
database : str, optional
Database name.
name_contains : str, optional
Select by a specific string on table name
name_prefix : str, optional
Select by a specific prefix on table name
name_suffix : str, optional
Select by a specific suffix on table name
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
Iterator[Dict[str, Any]]
Iterator of tables.
Examples
--------
>>> import awswrangler as wr
>>> tables = wr.catalog.get_tables()
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
paginator = client_glue.get_paginator("get_tables")
args: Dict[str, str] = {}
if catalog_id is not None:
args["CatalogId"] = catalog_id
if (name_prefix is not None) and (name_suffix is not None) and (name_contains is not None):
args["Expression"] = f"{name_prefix}.*{name_contains}.*{name_suffix}"
elif (name_prefix is not None) and (name_suffix is not None):
args["Expression"] = f"{name_prefix}.*{name_suffix}"
elif name_contains is not None:
args["Expression"] = f".*{name_contains}.*"
elif name_prefix is not None:
args["Expression"] = f"{name_prefix}.*"
elif name_suffix is not None:
args["Expression"] = f".*{name_suffix}"
if database is not None:
dbs: List[str] = [database]
else:
dbs = [x["Name"] for x in get_databases(catalog_id=catalog_id)]
for db in dbs:
args["DatabaseName"] = db
response_iterator = paginator.paginate(**args)
for page in response_iterator:
for tbl in page["TableList"]:
yield tbl
def tables(
limit: int = 100,
catalog_id: Optional[str] = None,
database: Optional[str] = None,
search_text: Optional[str] = None,
name_contains: Optional[str] = None,
name_prefix: Optional[str] = None,
name_suffix: Optional[str] = None,
boto3_session: Optional[boto3.Session] = None,
) -> pd.DataFrame:
"""Get a DataFrame with tables filtered by a search term, prefix, suffix.
Parameters
----------
limit : int, optional
Max number of tables to be returned.
catalog_id : str, optional
The ID of the Data Catalog from which to retrieve Databases.
If none is provided, the AWS account ID is used by default.
database : str, optional
Database name.
search_text : str, optional
Select only tables with the given string in table's properties.
name_contains : str, optional
Select by a specific string on table name
name_prefix : str, optional
Select by a specific prefix on table name
name_suffix : str, optional
Select by a specific suffix on table name
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
Iterator[Dict[str, Any]]
Pandas Dataframe filled by formatted infos.
Examples
--------
>>> import awswrangler as wr
>>> df_tables = wr.catalog.tables()
"""
if search_text is None:
table_iter = get_tables(
catalog_id=catalog_id,
database=database,
name_contains=name_contains,
name_prefix=name_prefix,
name_suffix=name_suffix,
boto3_session=boto3_session,
)
tbls: List[Dict[str, Any]] = list(itertools.islice(table_iter, limit))
else:
tbls = list(search_tables(text=search_text, catalog_id=catalog_id, boto3_session=boto3_session))
if database is not None:
tbls = [x for x in tbls if x["DatabaseName"] == database]
if name_contains is not None:
tbls = [x for x in tbls if name_contains in x["Name"]]
if name_prefix is not None:
tbls = [x for x in tbls if x["Name"].startswith(name_prefix)]
if name_suffix is not None:
tbls = [x for x in tbls if x["Name"].endswith(name_suffix)]
tbls = tbls[:limit]
df_dict: Dict[str, List] = {"Database": [], "Table": [], "Description": [], "Columns": [], "Partitions": []}
for table in tbls:
df_dict["Database"].append(table["DatabaseName"])
df_dict["Table"].append(table["Name"])
if "Description" in table:
df_dict["Description"].append(table["Description"])
else:
df_dict["Description"].append("")
df_dict["Columns"].append(", ".join([x["Name"] for x in table["StorageDescriptor"]["Columns"]]))
df_dict["Partitions"].append(", ".join([x["Name"] for x in table["PartitionKeys"]]))
return pd.DataFrame(data=df_dict)
def search_tables(text: str, catalog_id: Optional[str] = None, boto3_session: Optional[boto3.Session] = None):
"""Get Pandas DataFrame of tables filtered by a search string.
Parameters
----------
text : str, optional
Select only tables with the given string in table's properties.
catalog_id : str, optional
The ID of the Data Catalog from which to retrieve Databases.
If none is provided, the AWS account ID is used by default.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
Iterator[Dict[str, Any]]
Iterator of tables.
Examples
--------
>>> import awswrangler as wr
>>> df_tables = wr.catalog.search_tables(text='my_property')
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
args: Dict[str, Any] = {"SearchText": text}
if catalog_id is not None:
args["CatalogId"] = catalog_id
response: Dict[str, Any] = client_glue.search_tables(**args)
for tbl in response["TableList"]:
yield tbl
while "NextToken" in response: # pragma: no cover
args["NextToken"] = response["NextToken"]
response = client_glue.search_tables(**args)
for tbl in response["TableList"]:
yield tbl
def get_table_location(database: str, table: str, boto3_session: Optional[boto3.Session] = None) -> str:
"""Get table's location on Glue catalog.
Parameters
----------
database : str
Database name.
table : str
Table name.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
str
Table's location.
Examples
--------
>>> import awswrangler as wr
>>> wr.catalog.get_table_location(database='default', name='my_table')
's3://bucket/prefix/'
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
res: Dict[str, Any] = client_glue.get_table(DatabaseName=database, Name=table)
try:
return res["Table"]["StorageDescriptor"]["Location"]
except KeyError: # pragma: no cover
raise exceptions.InvalidTable(f"{database}.{table}")
def table(
database: str, table: str, catalog_id: Optional[str] = None, boto3_session: Optional[boto3.Session] = None
) -> pd.DataFrame:
"""Get table details as Pandas DataFrame.
Parameters
----------
database : str
Database name.
table : str
Table name.
catalog_id : str, optional
The ID of the Data Catalog from which to retrieve Databases.
If none is provided, the AWS account ID is used by default.
boto3_session : boto3.Session(), optional
Boto3 Session. The default boto3 session will be used if boto3_session receive None.
Returns
-------
pandas.DataFrame
Pandas DataFrame filled by formatted infos.
Examples
--------
>>> import awswrangler as wr
>>> df_table = wr.catalog.table(database='default', name='my_table')
"""
client_glue: boto3.client = _utils.client(service_name="glue", session=boto3_session)
if catalog_id is None:
tbl: Dict[str, Any] = client_glue.get_table(DatabaseName=database, Name=table)["Table"]
else:
tbl = client_glue.get_table(CatalogId=catalog_id, DatabaseName=database, Name=table)["Table"]
df_dict: Dict[str, List] = {"Column Name": [], "Type": [], "Partition": [], "Comment": []}
for col in tbl["StorageDescriptor"]["Columns"]:
df_dict["Column Name"].append(col["Name"])
df_dict["Type"].append(col["Type"])
df_dict["Partition"].append(False)
if "Comment" in col:
df_dict["Comment"].append(col["Comment"])
else:
df_dict["Comment"].append("")
for col in tbl["PartitionKeys"]:
df_dict["Column Name"].append(col["Name"])
df_dict["Type"].append(col["Type"])
df_dict["Partition"].append(True)
if "Comment" in col:
df_dict["Comment"].append(col["Comment"])
else:
df_dict["Comment"].append("")
return | pd.DataFrame(data=df_dict) | pandas.DataFrame |